binary.recode <- function(dat, pos.label = "Positive") { ## Purpose: Convert a data set to 0-1 format for binary diagnostic outcomes ## Arguments: ## dat: a data frame with binary diagnostic outcomes ## pos.label: a label for the positive outcome that is used in the "dat" ## Return: a data frame with binary diagnostic outcomes that are coded as numeric 0 or 1. ## Author: Feiming Chen ## ________________________________________________ dat[] <- lapply(dat, function(x, p=pos.label) ifelse(x == p, 1, 0)) dat } if (F) { # Unit Test binary.recode(data.frame(x = rep("Positive", 2), y = rep("Negative", 2))) ## x y ## 1 1 0 ## 2 1 0 }
Wednesday, November 13, 2019
Convert a data set to 0-1 format for binary diagnostic outcomes
Monday, August 5, 2019
Calculate Repeatability Coefficient (from Test-Retest Data)
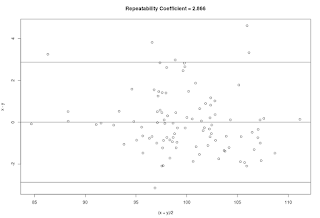
repeatability.coefficient <- function(x, y) { ## Purpose: Calculate Repeatability Coefficient (from Test-Retest Data) ## Arguments: ## x: First Test Outcome (continuous scale) ## y: Second Test Outcome (continuous scale) ## Return: Repeatability Coefficient ## Author: Feiming Chen ## ________________________________________________ N <- length(x) stopifnot(N == length(y)) s <- sqrt(sum((x - y)^2) / N) Repeatability.Coefficient <- 1.96 * s a <- paste("Repeatability Coefficient =", round(Repeatability.Coefficient, 3)) cat("N = ", N, "\n") cat("Mean Difference =", round(mean(x-y),3), "\n") cat("SD =", round(s, 3), "\n") cat(a, "\n") plot((x+y)/2, x-y, main = a) abline(h=c(0, -Repeatability.Coefficient, Repeatability.Coefficient)) } if (F) { # Unit Test N <- 100 u <- rnorm(N, mean = 100, sd = 5) x <- u + rnorm(N) y <- u + rnorm(N) repeatability.coefficient(x, y) # Theoretical True Value is 2 * sqrt(2) * 1 = 2.8284 ## Using ANOVA or Regression method for validation d <- data.frame(value = c(x, y), group = rep(1:N, 2)) aov(value ~ factor(group), data = d) # Residual standard error ~ 1 s <- summary(lm(value ~ factor(group), data = d))$sigma # Residual standard error ~ 1 ## Y1 - Y2 = E1 - E2, so Var(Y1-Y2) = Var(E1 - E2) = 2 * Var(E1) = 2 * s^2 cat("Repeatability Coefficient =", 1.96 * sqrt(2) * s, "\n") }
N = 100
Mean Difference = 0.012
SD = 1.433
Repeatability Coefficient = 2.866
Friday, June 28, 2019
Likelihood Ratios for Medical Decision Making
diagnostic.decision <- function(Sensitivity, Specificity, Prevalence = 0.01, AE.Risk = 0.005, Predicate.Sensitivity = 0.9, Predicate.Specificity = 0.9, Pretest.Prob = c(0.1, 0.25, 0.5, 0.75, 0.9), Cost.Benefit.Ratio = 1 ) { ## Purpose: Provide metrics to evaluate a medical test. ## Arguments: ## Sensitivity, Specificity: between 0 and 1. ## Prevalence: Disease Prevalence in the Target Population ## Pretest.Prob: a vector of pre-test probability ## AE.Risk: Risk of an adverse event due to treatment for False Positive patients. ## Predicate.Sensitivity, Predicate.Specificity: Performance of the comparator. ## Cost.Benefit.Ratio: Cost/Benefit = Cost(False Positive) / Cost(False Negative); default to 1. ## Return: Likelihood Ratios, Post-test Probability, Plots, etc. ## Author: Feiming Chen ## ________________________________________________ dev.close() p2o <- function(p) p/(1 - p) # convert probability to odds o2p <- function(o) o/(1 + o) # convert odds to probability r <- function(x) round(x, 2) # rounding function cat("\n----------- Diagnostic Performance for Subject Device ------------\n") cat("Sensitivity =", r(Sensitivity), "\n") cat("Specificity =", r(Specificity), "\n\n") cat("Risk Difference Evaluation:\n") J <- Sensitivity + Specificity - 1 # Risk Difference = Youden's Index = J cat("J =", r(J), "\n\n") LR.pos <- Sensitivity / (1 - Specificity) # Positive Likelihood Ratio (Risk Ratio) LR.neg <- (1 - Sensitivity) / Specificity # Negative Likelihood Ratio cat("Risk Ratio Evaluation:\n") cat("LR+ =", r(LR.pos), "\n") cat("LR- =", r(LR.neg), "\n\n") cat("Predictive Values:\n") PPV <- o2p(Sensitivity / (1 - Specificity) * p2o(Prevalence)) NPV <- o2p(Specificity / (1 - Sensitivity) / p2o(Prevalence)) cat("PPV = ", r(PPV), "\n") cat("NPV = ", r(NPV), "\n") cat("\nDecision Analysis:\n") cat("Cost/Benefit Ratio =", Cost.Benefit.Ratio, "\n") Risk.Threshold <- o2p(Cost.Benefit.Ratio) cat("Implied Risk Threshold =", r(Risk.Threshold), "\n") cat("\nRegret Graph Analysis:\n") abcissas.1 <- o2p(Cost.Benefit.Ratio / LR.pos) abcissas.2 <- o2p(Cost.Benefit.Ratio / LR.neg) cat("Disease Prevalence =", Prevalence, "\n") cat("Range of Disease Prevalence (Pre-Test Probability) to Choose a Test Strategy:\n") cat("0 --", r(abcissas.1), ": Test for None (Presume Negative; Treatment None)\n") cat(r(abcissas.1),"--", r(abcissas.2), ": Subject Device Test\n") cat(r(abcissas.2), "-- 1 : Test for All (Presume Positive; Treatment All)\n") cat("\n----------- Diagnostic Performance for Predicate Device ------------\n") cat("Sensitivity =", r(Predicate.Sensitivity), "\n") cat("Specificity =", r(Predicate.Specificity), "\n\n") cat("Risk Difference Evaluation:\n") J <- Predicate.Sensitivity + Predicate.Specificity - 1 # Risk Difference = Youden's Index = J cat("J =", r(J), "\n\n") LR2.pos <- Predicate.Sensitivity / (1 - Predicate.Specificity) # Positive Likelihood Ratio (Risk Ratio) LR2.neg <- (1 - Predicate.Sensitivity) / Predicate.Specificity # Negative Likelihood Ratio cat("Risk Ratio Evaluation:\n") cat("LR+ =", r(LR2.pos), "\n") cat("LR- =", r(LR2.neg), "\n\n") cat("\n----------- Compare the Subject and Predicate Device ------------\n") cat("Likelihood Ratio Graph:\n") plot(c(1 - Predicate.Specificity, 1 - Specificity), c(Predicate.Sensitivity, Sensitivity), xlim = c(0, 1), ylim = c(0, 1), asp = 1, xlab = "False Positive Rate", ylab = "True Positive Rate", pch = 19, col = c("red", "blue"), bty = "n", main = "Likelihood Ratio Graph") legend("bottomright", legend = c("Predicate", "Subject"), pch = 19, bg="lightyellow", col = c("red", "blue"), title = "Device") abline(0, LR.pos) min.spec <- 1 - 1/LR.pos cat("Minimum Specificity for a new test to be superior to the subject device in terms of PPV and NPV =", r(min.spec), "\n") min.sens <- 1 - LR.neg abline(min.sens, LR.neg) cat("Minimum Sensitivity for a new test to be superior to the subject device in terms of PPV and NPV =", r(min.sens), "\n") cat("\n----------- Expected Diagnostic Yield ------------\n") cat("Assume 1,000,000 patients from the target populaiton\n") cat("Assume the Disease Prevalence =", Prevalence, "\n") cat("Assume the Risk of an Adverse Event due to Treatment for False Positive Patients =", AE.Risk, "\n") cat("Predicate Device Performance: Sensitivity =", Predicate.Sensitivity, ", Specificity =", Predicate.Specificity, "\n") cat("Subject Device Performance: Sensitivity =", Sensitivity, ", Specificity =", Specificity, "\n\n") n <- 1000000 n1 <- round(n * Prevalence) # number with disease n2 <- n - n1 # number without disease EDY <- matrix(c(n1, n2, n1 * Sensitivity, n2 * (1 - Specificity), n1 * Predicate.Sensitivity, n2 * (1 - Predicate.Specificity), n1 * Sensitivity - n1 * Predicate.Sensitivity, n2 * (1 - Specificity) - n2 * (1 - Predicate.Specificity) ), 2) FP.per.TP <- apply(EDY, 2, function(x) round(x[2]/(x[1] + 0.01))) FP.per.TP.Pct <- FP.per.TP / FP.per.TP[1] * 100 EDY1 <- round(rbind(EDY, FP.per.TP, FP.per.TP.Pct)) dimnames(EDY1) <- list(c("Disease", "No.Disease", "Number.of.FP.Per.TP", "Percent.of.Worst.Case"), c("Expected.Number", "Subject+", "Predicate+", "Difference")) print(EDY1) cat("\nSafety Evaluation:\n") SAF <- matrix(round(c(n2, n2 * (1 - Specificity) * AE.Risk, n2 * (1 - Predicate.Specificity) * AE.Risk, n2 * (1 - Specificity) * AE.Risk - n2 * (1 - Predicate.Specificity) * AE.Risk, (n2 * (1 - Specificity) * AE.Risk - n2 * (1 - Predicate.Specificity) * AE.Risk) * 100 / (n1 * Sensitivity - n1 * Predicate.Sensitivity + 0.001))), 1, dimnames = list(c("No.Disease"), c("Expected.Number", "Subject+", "Predicate+", "Difference", "Number.of.AE.Per.100.Extra.TP"))) print(SAF) cat("\n----------- Medical Decision Making ------------\n") cat("Pre-test Probability =", Pretest.Prob, "\n") Pretest.Odds <- p2o(Pretest.Prob) cat("Pre-test Odds =", r(Pretest.Odds), "\n\n") Posttest.Odds.pos <- Pretest.Odds * LR.pos # with positive diagnostic result Posttest.Odds.neg <- Pretest.Odds * LR.neg # with negative diagnostic result Posttest.Prob.pos <- o2p(Posttest.Odds.pos) Posttest.Prob.neg <- o2p(Posttest.Odds.neg) cat("IF the diagnostic test is Positive:\n") cat("Post-test Odds =", r(Posttest.Odds.pos), "\n") cat("Post-test Probability =", r(Posttest.Prob.pos), "\n\n") cat("IF the diagnostic test is Negative:\n") cat("Post-test Odds =", r(Posttest.Odds.neg), "\n") cat("Post-test Probability =", r(Posttest.Prob.neg), "\n\n") N <- length(Pretest.Prob) x <- rep(0, N) dev.new() plot(c(-1.1, 1.1), c(0, 1), type="n", xlab="", ylab="Disease Probability", axes=F, main="Pre-Test Probability is updated to Post-Test Probability,\nafter a Diagnostic Test.", sub=paste("Diagnostic Test: Sensitivity =", r(Sensitivity), ", Specificity =", r(Specificity), ", LR+ =", r(LR.pos), ", LR- =", r(LR.neg))) arrows(x, Pretest.Prob, x-1, Posttest.Prob.neg, length=0.15) arrows(x, Pretest.Prob, x+1, Posttest.Prob.pos, length=0.15) points(x, Pretest.Prob, pch=19) points(x-1, Posttest.Prob.neg, pch=19) points(x+1, Posttest.Prob.pos, pch=19) axis(side=1, at=c(-1, 0, 1), labels=c("Post-Test (Test = NEG)", "Pre-Test", "Post-Test (Test = POS)")) axis(side=2, at=seq(0, 1, 0.1)) o <- 0.2 text(x, Pretest.Prob, labels=Pretest.Prob, pos=3, offset=o) text(x-1, Posttest.Prob.neg, labels=round(Posttest.Prob.neg, 2), pos=2, offset=o) text(x+1, Posttest.Prob.pos, labels=round(Posttest.Prob.pos, 2), pos=4, offset=o) list(EDY = EDY1, SAF = SAF) } if (F) { # Unit Test a <- diagnostic.decision(Sensitivity = 0.64, Specificity = 0.84, Prevalence = 0.0022, AE.Risk = 0.0068, Predicate.Sensitivity = 0.224, Predicate.Specificity = 0.881) }
----------- Diagnostic Performance for Subject Device ------------ Sensitivity = 0.64 Specificity = 0.84 Risk Difference Evaluation: J = 0.48 Risk Ratio Evaluation: LR+ = 4 LR- = 0.43 Predictive Values: PPV = 0.01 NPV = 1 Decision Analysis: Cost/Benefit Ratio = 1 Implied Risk Threshold = 0.5 Regret Graph Analysis: Disease Prevalence = 0.0022 Range of Disease Prevalence (Pre-Test Probability) to Choose a Test Strategy: 0 -- 0.2 : Test for None (Presume Negative; Treatment None) 0.2 -- 0.7 : Subject Device Test 0.7 -- 1 : Test for All (Presume Positive; Treatment All) ----------- Diagnostic Performance for Predicate Device ------------ Sensitivity = 0.22 Specificity = 0.88 Risk Difference Evaluation: J = 0.1 Risk Ratio Evaluation: LR+ = 1.88 LR- = 0.88 ----------- Compare the Subject and Predicate Device ------------ Likelihood Ratio Graph: Minimum Specificity for a new test to be superior to the subject device in terms of PPV and NPV = 0.75 Minimum Sensitivity for a new test to be superior to the subject device in terms of PPV and NPV = 0.57 ----------- Expected Diagnostic Yield ------------ Assume 1,000,000 patients from the target populaiton Assume the Disease Prevalence = 0.0022 Assume the Risk of an Adverse Event due to Treatment for False Positive Patients = 0.0068 Predicate Device Performance: Sensitivity = 0.224 , Specificity = 0.881 Subject Device Performance: Sensitivity = 0.64 , Specificity = 0.84 Expected.Number Subject+ Predicate+ Difference Disease 2200 1408 493 915 No.Disease 997800 159648 118738 40910 Number.of.FP.Per.TP 454 113 241 45 Percent.of.Worst.Case 100 25 53 10 Safety Evaluation: Expected.Number Subject+ Predicate+ Difference Number.of.AE.Per.100.Extra.TP No.Disease 997800 1086 807 278 30 ----------- Medical Decision Making ------------ Pre-test Probability = 0.1 0.25 0.5 0.75 0.9 Pre-test Odds = 0.11 0.33 1 3 9 IF the diagnostic test is Positive: Post-test Odds = 0.44 1.33 4 12 36 Post-test Probability = 0.31 0.57 0.8 0.92 0.97 IF the diagnostic test is Negative: Post-test Odds = 0.05 0.14 0.43 1.29 3.86 Post-test Probability = 0.05 0.12 0.3 0.56 0.79
Tuesday, March 5, 2019
Bland Altman Limit of Agreement Calculation with the presence of repeated measurements where the true value vary
Refer to this blog post for the function var.comp.one.way.model
bland.altman.repeated.vary <- function(fmla, dat) { ## Purpose: Calculate LoA (Limit of Agreement) when data has repeated measurements (true value varies) ## Arguments: ## fmla: something like "Difference ~ Subject". ## dat: data frame with "Difference" and "Subject". ## Return: LoA ## Author: Feiming Chen ## Reference: ## Bland, J. Martin, and Douglas G. Altman. "Agreement between methods of measurement with multiple observations per ## individual." Journal of biopharmaceutical statistics 17.4 (2007): 571-582. ## ________________________________________________ ans <- var.comp.one.way.model(fmla, dat) s <- ans$VCA$SD[3] m <- ans$Mean cat("\nMean =", m, ", SD =", s, "\n") loa <- c(m - 1.96*s, m + 1.96*s) cat("\n95% Limits of Agreement: (", loa[1], ",", loa[2], ")\n") invisible(list(m=m, s=s, loa=loa)) } if (F) { # Unit Test dat <- data.frame(Subject = factor(c(1, 1, 1, 2, 2, 2, 3, 3, 3, 3)), A = rnorm(10, 10), B = rnorm(10, 10)) dat$Difference <- dat$A - dat$B fmla <- Difference ~ Subject bland.altman.repeated.vary(fmla, dat) ## Mean = 0.0997215 , SD = 1.851 ## 95% Limits of Agreement: ( -3.528239 , 3.727681 ) }
Subscribe to:
Posts (Atom)


