compare.tables <- function(d1, d2, by.idx = 1)
{
## Purpose: Compare two tables with the same structure by merging them.
## Arguments:
## d1: table A
## d2: table B
## by.idx: an index vector to common variables.
## Return: a full joined table for comparision
b <- names(d1)[by.idx]
d <- full_join(d1, d2, by = b)
tex.print(d, type = "HTML")
invisible(d)
}
if (F) { # Unit Test
d1 <- rc()
d2 <- rc()
by.idx <- 1:4
compare.tables(d1, d2, by.idx)
}
Tuesday, December 12, 2017
Compare two tables with the same structure by merging them
List duplicated files in a directory
list.dup.files <- function(p, file.name.head = 20)
{
## Purpose: List duplicated files in a directory.
## Arguments:
## file.name.head: number of starting characters in the file name to use for duplication detection
## Return: a list of file names that are duplicated (by sorted order).
## Author: Feiming Chen, Date: 13 Nov 2017, 16:36
## ________________________________________________
p1 <- dir(p)
p2 <- sort(p1, decreasing = T)
p3 <- as.character(sapply(p2, function(x) substr(x, 1, file.name.head)))
p4 <- p2[duplicated(p3)]
if (length(p4) > 0) {
cat("Duplicated Older Files:\n")
print(p4)
} else {
cat("NO Duplicated Files.\n")
}
file.path(p, p4)
}
if (F) { # Unit Test
p <- "~/tmp/Updated-Databases"
file.name.head <- 25
x <- list.dup.files(p, file.name.head)
file.remove(x)
}
Wednesday, October 18, 2017
Convert all character variables in your data.frame to factors
df.make.factors <- function(df)
{
## Purpose: Convert all character variables in your data.frame to factors
## Arguments:
## df: a data frame
## Return: a data frame with converted factors on those character variables.
## Author: Feiming Chen, Date: 17 Oct 2017, 14:26
## ________________________________________________
character_vars <- sapply(df, class) == "character"
df[, character_vars] <- lapply(df[, character_vars], factor)
df
}
if (F) { # Unit Test
df <- data.frame(x=c("x", "x", "y"), y=1:3, z=c("1", "3", "3"))
df.make.factors(df)
}
Monday, October 2, 2017
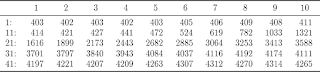
Display a (numeric) vector in a pretty format
display.vector <- function(x)
{
## Purpose: Display a vector in a pretty format
## Arguments:
## x: a vector
## Return: a png picture for the vector. (each row has 10 elements)
## Author: Feiming Chen, Date: 2 Oct 2017, 14:54
## ________________________________________________
N <- length(x)
n1 <- N %% 10
if (n1 > 0) {
n2 <- 10 - n1
x.add <- rep(NA, n2)
x <- c(x, x.add)
}
x1 <- matrix(x, ncol = 10, byrow = T)
n2 <- nrow(x1)
nr <- paste0(10 * (seq.int(n2) - 1) + 1, ": ")
dimnames(x1) <- list(nr, 1:10)
tex.print(x1)
}
if (F) { # Unit Test
x <- c(402.6615584,402.1343365,402.5467634,401.835917,402.9268321,404.5185276,406.0735012,408.7713965,408.2033098,411.043676,413.953578,420.6021333,426.6352944,440.9007592,471.5614004,523.8426555,619.173643,782.3175971,1033.289048,1321.267143,1615.935774,1898.979548,2173.03472,2442.864835,2682.25644,2884.575978,3064.278857,3252.58248,3413.084683,3588.447246,3701.244129,3796.824264,3840.251429,3943.184319,4084.131835,4037.30822,4116.338229,4191.71438,4173.699815,4110.847391,4197.181334,4221.081422,4207.343855,4209.140892,4263.463393,4306.872737,4312.087451,4269.522631,4313.924265,4265.306511)
display.vector(x)
}
tex.print <- function(x, type=c("PNG", "HTML"), file=NULL, caption = file, digits=0, ...) {
## Purpose: Generate HTML or PNG representation of a data/table/model object.
## Require R "memisc" package.
## Arguments:
## x: an object (table, ftable, data frame, model object, etc. that are acceptable in R "memisc" package.
## type: what to generate --
## "HTML" shows a HTML table in a web browser.
## "PNG" shows a PNG (image) file (require linux commands: latex, dvipng, display).
## file: make a copy of the PNG file with this name. Also used for the table name.
## caption: table caption.
## digits: number of decimal places to use.
## ...: passed to "toLatex"
## Return: Display the data representation and an image file (type="PNG") for insertion.
## Author: Feiming Chen, Date: 14 Feb 2017, 11:59
## ________________________________________________
require(memisc)
type <- match.arg(type)
switch(type,
HTML = {
## Show HTML table in a Web Browser
memisc::show_html(x)
},
PNG = {
## Generate LaTeX and PNG Image file
r <- toLatex(x, ..., digits=digits, show.vars=TRUE)
f <- file("X.tex", "w")
writeLines(c("\\documentclass{article}",
"\\usepackage{booktabs}",
"\\usepackage{dcolumn}",
"\\begin{document}",
"\\pagenumbering{gobble}"), f)
if (!is.null(file)) writeLines("\\begin{table} \\centering", f)
writeLines(r, f)
if (!is.null(caption)) {
writeLines(paste0("\\caption{", caption, "} \\end{table}"), f)
}
writeLines("\\end{document}", f)
close(f)
system("latex X.tex >/dev/null") # Generate DVI file from LaTeX file (X.tex -=> X.dvi)
system("dvipng -q* -o X.png -T tight -D 200 X.dvi >/dev/null") # convert to PNG file with Tight box and Resolution of 200
system("display X.png &") # display the PNG file.
file.remove("X.tex", "X.log", "X.aux", "X.dvi")
if (!is.null(file)) file.copy("X.png", paste0(file,".png"), overwrite = T)
if (!grepl("/tmp", getwd())) {
file.copy("X.png", "~/tmp/X.png", overwrite = T)
}
}
)
invisible(NULL)
}
Thursday, September 7, 2017
Extracts all calls to a function (except for those in standard packages)
my.fun_calls <- function(f)
{
## Purpose: Extracts all calls to a function (except for those in standard packages) in a function
## Differs from pryr::fun_calls by excluding basic functions.
## Arguments:
## f: a function
## Return: All calls to a function (that are not in standard packages, e.g. base, stats, etc.)
## Author: Feiming Chen, Date: 7 Sep 2017, 11:23
## ________________________________________________
if (is.function(f)) {
my.fun_calls(body(f))
} else if (is.call(f)) {
fname <- as.character(f[[1]])
if (identical(fname, ".Internal")) fname3 <- NULL
f1 <- try(pryr::where(fname), silent = TRUE)
if (is.tryerror(f1)) {
if (fname[1] == "::") { # Example: pryr::where fname = c("::", "pryr", "where")
fname3 <- paste0(fname[2], fname[1], fname[3])
} else {
fname3 <- NULL
}
} else {
fname2 <- environmentName(f1)
if (fname2 %in% c("base", "package:stats", "package:graphics", "package:grDevices", "package:utils", "package:datasets", "package:methods")) {
fname3 <- NULL
} else {
if (fname2 == "R_GlobalEnv") {
fname3 <- fname
} else { # attach package name if other than Global Environment
fname2m <- sub("package:", "", fname2) # remove "package:" prefix.
if (fname2m == "") {
fname3 <- paste0(fname)
} else {
fname3 <- paste0(fname2m, "::", fname)
}
}
}
}
if (F) fun_calls(lm) # for testing purpose only
unique(c(fname3, unlist(lapply(f[-1], my.fun_calls), use.names = FALSE)))
}
}
if (F) { # Unit Test
my.fun_calls(lm) # NULL
sort(my.fun_calls(my.fun_calls))
## "is.tryerror" "my.fun_calls" "pryr::fun_calls" "pryr::where"
}
## Test for the try-error class (from "try" function)
is.tryerror <- function(x) inherits(x, "try-error")
Wednesday, June 28, 2017
Span: A Predicate Functional that finds the location of the longest sequential run of elements where the predicate is true
Span <- function(f, x)
{
## Purpose: A Predicate Functional that finds the location of the longest sequential run
## of elements where the predicate is true.
## Arguments:
## f: a predicate (a function that returns a single TRUE or FALSE)
## x: a list or data frame
## Return: The location of the longest sequential run of elements where the predicate is true.
## Author: Feiming Chen, Date: 28 Jun 2017, 09:45
## ________________________________________________
y <- rle(Where(f, x))
z <- y$lengths
w <- which.max(z) # position of the segment with longest sequential run of TRUE
pos.end <- sum(z[1:w])
pos.start <- pos.end - z[w] + 1
c(start=pos.start, end=pos.end)
}
if (F) { # Unit Test
x <- c(3, 1, NA, 4, NA, NA, NA, 5, 9, NA, NA)
Span(is.na, x)
## start end
## 5 7
}
Where <- function(f, x)
{
## Purpose: A Predicate Functional
## See other Predicate Functionals: Filter(), Find(), Position()
## See other Functionals: Map(), Reduce()
## Arguments:
## f: a predicate (a function that returns a single TRUE or FALSE) (e.g. is.character, all, is.NULL)
## x: a list or data frame
## Return: a logical vector
## Author: Feiming Chen, Date: 28 Jun 2017, 09:45
## ________________________________________________
vapply(x, f, logical(1))
}
if (F) { # Unit Test
df <- data.frame(x = 1:3, y = c("a", "b", "c"), stringsAsFactors = T)
Where(is.factor, df)
## Compare with:
Filter(is.factor, df)
Find(is.factor, df)
Position(is.factor, df)
}
Thursday, June 22, 2017
Draw a data frame as a R plot
my.table.plot0 <- function(x,
highlight.matrix = NULL,
main = "Table", title2 = NULL, size = 20,
bold.lastrow = FALSE, bold.lastcol = FALSE
)
{
## PURPOSE: Draw a data frame as a R plot. Require R package "gridExtra".
## ARGUMENT:
## x: a data frame
## highlight.matrix: if provided a logical matrix (same dimension as "x"). Then "TRUE" cells will be highlighted.
## main: title for the plot
## title2: additional title with more verbose text
## size: text size of table text
## bold.lastrow: if T, make the last row bold font.
## bold.lastcol: if T, make the last column bold font.
## RETURN: a plot
## DATE: 21 Nov 2016, 15:46
## -----------------------------------------------------
x <- as.data.frame(x)
n1 <- nrow(x)
n2 <- ncol(x)
cl <- matrix("black", n1, n2)
cl[is.na(x)] <- "gray"
## Bold Total Column and Row
cl2 <- cl
cl2[] <- "plain"
if (bold.lastrow) {
cl2[n1,] <- "bold"
}
if (bold.lastcol) {
cl2[, n2] <- "bold"
}
require(gridExtra)
table_theme <- ttheme_default(base_size = size,
core = list(fg_params=list(col=cl, fontface = cl2)))
## Highlight Cells
if (!is.null(highlight.matrix)) {
cl3 <- cl
z <- ttheme_default()$core$bg_params$fill # default color
z1 <- rep(z, length.out = nrow(cl3))
for (i in 1:ncol(cl3)) cl3[,i] <- z1
cl3[highlight.matrix] <- "yellow"
table_theme$core$bg_params$fill <- cl3
}
plot.new()
gridExtra::grid.table(as.data.frame(x), theme=table_theme)
## Title Text
t2 <- ifelse(is.null(title2), "", paste0("\n", graph.title.split.lines(title2, sep=" ", num.chars = 40)))
title(main = paste0(main, t2))
}
if (F) { # Unit Test
x <- data.frame(Type=c('Car', 'Plane'), Speed=c(NA, 200))
my.table.plot0(x)
my.table.plot0(x, main='Test', title2='a very complex test', size=30, bold.lastcol = T, highlight.matrix = matrix(c(NA, F, F, T) , 2, 2))
x <- data.frame(Type=rep('Car', 20), Speed=rep(100, 20))
my.table.plot0(x)
}
my.table.plot <- function(x, highlight.matrix = NULL, bold.lastrow = FALSE, n = 20, ...)
{
## Purpose: Split a data frame to fit in each page and call "my.table.plot0".
## Arguments:
## x: a data frame or matrix
## highlight.matrix: if provided a logical matrix (same dimension as "x"). Then "TRUE" cells will be highlighted.
## bold.lastrow: if T, make the last row bold font.
## n: how many rows to fit into each page.
## ...: pass to "my.table.plot0".
## Return: multiple plots to plot the data matrix.
## Author: Feiming Chen, Date: 5 Jul 2017, 15:16
## ________________________________________________
x <- as.data.frame(x)
N <- nrow(x)
if ( N <= n ) {
my.table.plot0(x, highlight.matrix = highlight.matrix, bold.lastrow = bold.lastrow, ...)
} else {
s <- rep(seq.int(N %/% 20 + 1), each = n, length.out = N)
xs <- split(x, s)
ng <- length(xs) # number of pages
bold.lastrow.list <- c(rep(FALSE, ng-1), bold.lastrow)
if (is.null(highlight.matrix)) {
mapply(my.table.plot0, xs, bold.lastrow = bold.lastrow.list, MoreArgs = list(...))
} else {
ys <- split(highlight.matrix, s)
mapply(my.table.plot0, xs, highlight.matrix = ys, bold.lastrow = bold.lastrow.list, MoreArgs = list(...))
}
}
}
if (F) { # Unit Test
x <- data.frame(Type=rep('Car', 50), Speed=rep(100, 50))
h <- matrix(rep(c(F, T), 50), ncol=2)
my.table.plot(x, highlight.matrix = h, bold.lastrow = T)
}
Wednesday, June 21, 2017
Add a "Total" level to each specified categorical variable in the data frame
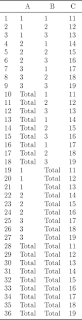
df.add.total.category <- function(dat, var.list, new.level.label = "Total")
{
## Purpose: Add a "Total" level to each specified categorical variable in the data frame.
## Arguments:
## dat: a data frame with several categorical variables for making tables.
## var.list: a list of variable names in "dat".
## new.level.label: the name for the new "total" level for the specified variable.
## Return:
## Author: Feiming Chen, Date: 21 Jun 2017, 13:02
## ________________________________________________
if ( missing(var.list) ) {
var.list <- names(dat)
}
d0 <- dat # to make new label across all variables
lapply(var.list, function( v ) {
d <- dat
d[[v]] <- new.level.label
d0[[v]] <<- new.level.label
d
}) -> dd
do.call(rbind, c(list(dat), dd, list(d0)))
}
if (F) { # Unit Test
dat <- data.frame(A=gl(3,3), B=gl(3, 1, 9), C=11:19)
df.add.total.category(dat, var.list=c("A", "B"))
}
Monday, June 19, 2017
Put unique elements of a string vector into one long string
paste.unique.string <- function(x, sep = "/")
{
## Purpose: Put unique elements of a string vector into one long string.
## Arguments:
## x: a string vector
## sep: what separator to use when pasting all unique elements together.
## Return: a single string with unique elements of "x".
## Author: Feiming Chen, Date: 19 Jun 2017, 14:00
## ________________________________________________
paste0(unique(x), collapse = sep)
}
if (F) { # Unit Test
paste.unique.string(c("a", "b", "a", "b")) # "a/b"
}
Add "0" to the front of numbers to make n-digit string
prepend.zero.to.numbers <- function(x, n = NULL, pad.char = "0")
{
## Purpose: Add "0" to the front of numbers to make n-digit string.
## Arguments:
## x: a vector of integers/characters
## n: Number of total digits after padding with zero. Default to
## using the largest number to calculate "n".
## pad.char: what character to pad. Default to "0".
## Return: a character vector with nuumbers padded with zero's in the head.
## Author: Feiming Chen, Date: 19 Jun 2017, 13:19
## ________________________________________________
x1 <- as.character(x)
y <- nchar(x1)
if (is.null(n)) n <- nchar(as.character(max(x)))
y1 <- n - y # how long is the padding for each number
pad.vector <- sapply(y1, function(w) paste(rep(pad.char, w), collapse = ""))
paste0(pad.vector, x1)
}
if (F) { # Unit Test
x <- c(2, 14, 156, 1892)
prepend.zero.to.numbers(x) # "0002" "0014" "0156" "1892"
prepend.zero.to.numbers(x, pad.char = "X") # "XXX2" "XX14" "X156" "1892"
prepend.zero.to.numbers(x, n = 6) # "000002" "000014" "000156" "001892"
y <- c("A", "BC", "DEF")
prepend.zero.to.numbers(y, 3) # "00A" "0BC" "DEF"
}
Parse a string to generate a data frame
parse2data.frame <- function(x) { ## Purpose: Parse a string to generate a data frame. ## Arguments: ## x: a string with header and multiple rows of text that resembles a data frame. ## Fields are separated by comma. ## Return: a data frame ## Author: Feiming Chen, Date: 19 Jun 2017, 10:12 ## ________________________________________________ d <- read.csv(textConnection(x), as.is = T) i <- sapply(d, is.character) d[i] <- lapply(d[i], trimws) d } if (F) { # Unit Test a = " Test, ID, Name t1 , 3, Sun t2 , 5, Moon t3, 2, Earth " b <- parse2data.frame(a) str(b) ## 'data.frame': 3 obs. of 2 variables: ## $ ID : int 3 5 2 ## $ Name: Factor w/ 3 levels " Earth"," Moon",..: 3 2 1 }
Thursday, June 15, 2017
Tokenize a string into a vector of tokens
tokenize.string <- function(x, split = "[ ,:;]+")
{
## Purpose: Tokenize a string into a vector of tokens
## Arguments:
## x: a string or a vector of strings
## split: split characters (regular expression)
## Return: a character vector (if "x" is a string) or a list of character vectors.
## Author: Feiming Chen, Date: 15 Jun 2017, 15:01
## ________________________________________________
ans <- strsplit(x, split="[ ,:;]+", fixed=F)
if (length(x) == 1) ans <- ans[[1]]
ans
}
if (F) { # Unit Test
x <- "IND, UNR INC ; TCU"
tokenize.string(x)
## [1] "IND" "UNR" "INC" "TCU"
tokenize.string(rep(x, 3))
## [[1]]
## [1] "IND" "UNR" "INC" "TCU"
## [[2]]
## [1] "IND" "UNR" "INC" "TCU"
## [[3]]
## [1] "IND" "UNR" "INC" "TCU"
}
Wednesday, June 14, 2017
Count the number of distinct values in a vector or distinct rows in a data frame
N.levels <- function(x)
{
## Purpose: Count the number of distinct values in a vector or distinct rows in a data frame
## Arguments:
## x: a vector (numeric or string), or a data frame.
## Return: a count for the distinct values/rows in the vector or data frame.
## Author: Feiming Chen, Date: 20 Mar 2017, 10:58
## ________________________________________________
sum(!duplicated(x))
}
if (F) { # Unit Test
N.levels(c(1,2,1,3)) # 3
N.levels(c("a", "b", "a")) # 2
x <- data.frame(a=c(1,2,1), b=c(1, 2, 1))
N.levels(x) # 2
}
Tuesday, June 6, 2017
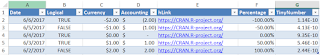
Write data tables to Excel (.xlsx) file
wxls <- function(x, file = "test", ...)
{
## Purpose: Write data tables to Excel (.xlsx) file.
## Require package "openxlsx".
## Arguments:
## x: a data frame or a list of data frames.
## file: a naked file name with no extension.
## ...: passed to "write.xlsx".
## Return: Generate an Excel (.xlsx) file.
## Author: Feiming Chen, Date: 6 Jun 2017, 15:11
## ________________________________________________
require(openxlsx)
f <- paste0(file, ".xlsx")
write.xlsx(x, file = f, asTable = TRUE,
creator = "Feiming Chen",
tableStyle = "TableStyleMedium2", ...)
}
if (F) { # Unit Test
df <- data.frame("Date" = Sys.Date()-0:4,
"Logical" = c(TRUE, FALSE, TRUE, TRUE, FALSE),
"Currency" = paste("$",-2:2),
"Accounting" = -2:2,
"hLink" = "https://CRAN.R-project.org/",
"Percentage" = seq(-1, 1, length.out=5),
"TinyNumber" = runif(5) / 1E9, stringsAsFactors = FALSE)
class(df$Currency) <- "currency"
class(df$Accounting) <- "accounting"
class(df$hLink) <- "hyperlink"
class(df$Percentage) <- "percentage"
class(df$TinyNumber) <- "scientific"
wxls(df)
wxls(list(A = df, B = df, C= df)) # write to 3 separate tabs with corresponding list names.
}
Monday, June 5, 2017
Plot "y" against "x" where "x" is a vector of labels
my.plot.vs.label <- function(x, y, ...)
{
## Purpose: Plot "y" against "x" where "x" is a vector of labels (string)
## Arguments:
## x: a string vector (labels)
## y: a numeric vector or matrix
## ...: passed to "matplot"
## Return: a plot
## Author: Feiming Chen, Date: 5 Jun 2017, 14:46
## ________________________________________________
s <- seq(x)
matplot(s, y, type="b", axes = F, xlab ="", ylab="", ...)
axis(side=2)
axis(side=1, at=s, labels=x, las=2)
yn <- ncol(y)
yna <- names(y)
if (!is.null(yn) && !is.null(yna)) {
legend("topright", legend=yna, col=1:yn, lwd=1.5, lty=1:yn)
}
}
if (F) { # Unit Test
x <- LETTERS
y <- matrix(rnorm(260), ncol=10)
my.plot.vs.label(x, y)
y2 <- as.data.frame(y)
my.plot.vs.label(x, y2)
}
Friday, June 2, 2017
Read/Import all CSV files in a specified directory
batch.read.csv.files <- function(p)
{
## Purpose: Read/Import all CSV files in a specified directory.
## Require package "readr".
## Arguments:
## p: a file directory with CSV files.
## Return:
## a list, each element is a data frame (imported from the file) and its name is the corresponding file name.
## Author: Feiming Chen, Date: 2 Jun 2017, 11:09
## ________________________________________________
file.list <- list.files(p, pattern=".csv", full.names = TRUE, ignore.case = TRUE)
if (length(file.list) > 0) {
dat <- lapply(file.list, readr::read_csv)
names(dat) <- sapply(file.list, basename)
} else dat <- list()
cat("Import", length(dat), "CSV Files.\n")
dat
}
Wednesday, May 3, 2017
Feature Filtering with Natural B-Splines for Functional Modeling Application
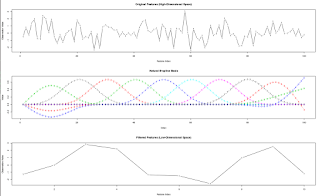
feature.filtering.ns <- function(X, df = 10, graph = FALSE)
{
## Purpose: Feature Filtering with Natural B-Splines for Functional Modeling Application.
## Preprocess the feature data (design matrix) to enforce a natural regularization.
## In other words, project each observation to each of "df" axis represented by B-spline Basis.
## Arguments:
## X: a design matrix (predictors) of dimension N x p (N observations, p predictors).
## If "X" is a vector, convert it to a matrix with one row.
## df: Degrees of Freedom (number of natural B-splines basis functions to approximate the (continuous) coefficient function.
## Beta (p x 1) = H (p x df) Theta (df x 1), where H represents "df" number of continuous basis functions
## in the coefficient space, and Theta represents the linear combination parameters for constructing Beta.
## graph: if TRUE, plot each observation (each row of X) as a time series,
## its projection to the reduced space (formed by the natural B-spline basis),
## and the natural B-spline basis.
## Return: Preprocessed Features (X H) of dimension N x df, which can be used as "df" input into a predictive model.
## X Beta = X H Theta = (X H) Theta
## Author: Feiming Chen, Date: 3 May 2017, 12:12
## ________________________________________________
if (is.vector(X)) X <- matrix(X, nrow = 1)
p <- ncol(X) # number of predictors
require(splines)
## B-splines for Natural Cubic Spline: a p x df matrix.
H <- ns(1:p, df=df)
## Filtered Features. Dimensions: N x p p x df = N x df
## Project each observation "x" into one of "df" Basis (each column of H matrix) so that
## each "x" is represented by "df" coefficient (coordinates in the coordinate system defined by "H")
ans <- X %*% H
if (graph) {
par(mfrow=c(3,1))
matplot(t(X), type = "b", xlab = "Feature Index", ylab = "Observation Value",
main = "Original Features (High-Dimensional Space)")
matplot(H, type="b", xlab = "Index", ylab ="Value", main="Natural B-spline Basis")
matplot(t(ans), type = "b", xlab = "Feature Index", ylab = "Observation Value",
main = "Filtered Features (Low-Dimensional Space)")
par(mfrow=c(1,1))
}
ans
}
if (F) { # Unit Test
X <- matrix(rnorm(1000), 20, 50)
y <- feature.filtering.ns(X)
y <- feature.filtering.ns(X, df = 20)
x <- rnorm(100)
y <- feature.filtering.ns(x, graph=T)
}
Tuesday, April 25, 2017
Apply a stamp (text string) to each page of a PDF file
my.pdf.stamp <- function(stamp.text="Stamp", pos=c(0.4, 0.9), pdf.in.file, pdf.out.file)
{
## Purpose: Apply a stamp (text string) to each page of a PDF file.
## Requires Linux program "pdftk".
## Arguments:
## pdf.in.file: Input PDF file.
## stamp.text: a vector of strings (stamps). Each value will be stamped on the corresponding page of the PDF file.
## Its values will be recycled to match the total number of pages in the input PDF file.
## pos: position of the stamp text (located between [0,1] X [0,1])
## pdf.out.file: Output PDF file with the stamps.
## Return: PDF file with the stamps.
## Author: Feiming Chen, Date: 25 Apr 2017, 10:42
## ________________________________________________
p1 <- pdf.file.page.number(pdf.in.file)
p2 <- length(stamp.text)
if (p1 != p2) stamp.text <- rep(stamp.text, length.out = p1)
## burst the PDF file into single page PDF's
f <- "page_%05d.pdf"
fs <- "stamp_%05d"
fs2 <- "stamp_%05d-Plot.pdf"
fo <- "out_%05d.pdf"
system(paste("pdftk", pdf.in.file, "burst output", f))
for (i in 1:p1) {
X(text.in.plot(stamp.text[i], cex = 2, pos = pos), file = sprintf(fs, i), open.pdf=F)
system(paste("pdftk", sprintf(f, i), "stamp", sprintf(fs2, i), "output", sprintf(fo, i)))
}
merge.file.list <- paste(sapply(1:p1, function(i) sprintf(fo, i)), collapse = " ")
system(paste("pdftk", merge.file.list, "cat output", pdf.out.file))
}
if (F) { # Unit Test
dir.create("tmp"); setwd("tmp")
pdf("test.pdf"); replicate(3, plot(rnorm(100))); dev.off()
pdf.in.file <- "test.pdf"
pdf.out.file <- "result.pdf"
stamp.text <- LETTERS[1:3]
pos <- c(0.4, 0.9)
my.pdf.stamp(stamp.text, pos=pos, pdf.in.file, pdf.out.file)
}
pdf.file.page.number <- function(fname) {
## Return the total number of pages in a PDF file.
## Require Linux program "pdfinfo"
a <- pipe(paste0("pdfinfo '", fname, "' | grep Pages | cut -d: -f2"))
page.number <- as.numeric(readLines(a))
close(a)
page.number
}
if (F) {
## pdf.file.page.number("ALL-Plots.pdf")
}
text.in.plot <- function(x, cex=1, pos=c(0.5, 0.5)) {
## "x" is a string. Put "x" in the center of a plot. For putting
## text summary in a PDF file that is mostly plots.
## "pos" is the position of the text string
## "cex" is the expansion ratio of the text string
plot(c(0, 1), c(0, 1), type = "n", main = "", xlab = "", ylab = "", xaxt="n", yaxt="n", axes = F, mar=c(0,0,0,0), oma=c(0,0,0,0))
text(pos[1], pos[2], x, cex=cex)
}
if (F) {
text.in.plot("good", cex=5)
text.in.plot("bad", pos=c(0.1, 0.1))
}
X <- function(expr, file="test", open.pdf = TRUE) {
## USED IN UNIX!
## Generate a PDF file based on evaluation of expr and open it
## Put multiple plot commands like "{ cmd1; cmd2; cmd3; cmd4 }".
## USAGE: xpdf(plot(sig1))
graphics.off()
## png("~/tmp/plot.png", width=600, height=600)
r <- my.pdf(file)
ans <- eval(expr)
dev.off()
## system("convert ~/tmp/plot.pdf ~/tmp/plot.png")
if (open.pdf) open.pdf(r$plot)
invisible(ans)
}
if (F) {
X(plot(rnorm(100)))
a <- X({plot(a <- rnorm(100)); a})
X({plot(a <- rnorm(100)); hist(a); boxplot(a); dotchart(a); qqnorm(a); plot(lm(a~seq(a)))})
}
open.pdf <- function(file) {
## Open a PDF file.
system(paste("evince ", file, " &", sep=""))
cat("See Plot In: ", file, "\n")
}
Return the total number of pages in a PDF file.
pdf.file.page.number <- function(fname) {
## Return the total number of pages in a PDF file.
## Require Linux program "pdfinfo"
a <- pipe(paste0("pdfinfo '", fname, "' | grep Pages | cut -d: -f2"))
page.number <- as.numeric(readLines(a))
close(a)
page.number
}
if (F) {
## pdf.file.page.number("ALL-Plots.pdf")
}
Friday, April 21, 2017
Rename all files in a directory according to a pattern (regular expression)
batch.rename.file <- function(pattern, replacement, path=".")
{
## Purpose: Rename all files in a directory according to a pattern (regular expression).
## Arguments:
## pattern: a regular expression (as is used by R base function "sub") to match the file names.
## replacement: replacement text for the new file name (as is used by R base function "gsub")
## path: path to a directory containing the files to be renamed. Default to the current directory.
## Return: Side Effect (all files in the directory will be renamed)
## Author: Feiming Chen, Date: 21 Apr 2017, 13:32
## ________________________________________________
f1 <- dir(path, pattern, full.names = T, recursive = T) # Current file names. Include all files in subdirectories.
f1.base <- basename(f1)
f2.base <- sub(pattern, replacement, f1.base) # Corresponding New file names
f2 <- file.path(dirname(f1), f2.base)
ans <- file.rename(f1, f2)
idx <- which(!ans) # which operation (file renaming) fails
if (length(idx) == 0) {
cat("All", length(ans), "files are renamed.\n")
} else {
cat(length(idx), "out of", length(ans), "files are not renamed.\n")
}
}
if (F) { # Unit Test
dir.create("tmp")
setwd("tmp")
file.create("a.b.html", "c.d.html")
batch.rename.file("\\.", "_")
dir() # "a_b.html" "c_d.html"
}
Thursday, April 20, 2017
Abbreivate a binomial scientific name
abbreviate.species.names <- function(x)
{
## Purpose: Abbreivate a binomial scientific name.
## Arguments:
## x: a vector of strings for the names of the species.
## Return: a vector of strings with the abbreviated names
## Author: Feiming Chen, Date: 20 Apr 2017, 14:01
## ________________________________________________
x1 <- gsub(" +", " ", x)
x2 <- sub("^ +", "", x1)
x3 <- sub(" +$", "", x2)
x4 <- strsplit(x3, " ")
sapply(x4, function(y) {
if (length(y) == 2) {
ans <- paste0(toupper(substr(y[1], 1, 1)), ".", tolower(y[2]))
} else ans <- y
ans
})
}
if (F) { # Unit Test
x = c("Escherichia coli", "Tyrannosaurus rex", "N/A", " Canis lupus ")
abbreviate.species.names(x) # "E.coli" "T.rex" "N/A" "C.lupus"
}
Tuesday, April 18, 2017
Extract a specific "pattern" from each element of a character vector
extract.pattern <- function(x, pattern = "([[:alnum:]]+)")
{
## Purpose: Extract a specific "pattern" from each element of a character vector.
## Arguments:
## x: a character vector
## pattern: a regular expression which specifies the pattern (inside brackets) to be extracted.
## Return: a vector with the extracted pattern. NA is filled in where no match is found.
## Author: Feiming Chen, Date: 18 Apr 2017, 14:34
## ________________________________________________
r <- paste0(".*", pattern, ".*")
sub(r, "\\1", x)
}
if (F) { # Unit Test
x = c(NA, "a-b", "a-d", "b-c", "d-e")
extract.pattern(x) # extract alpha-numeric pattern
## [1] NA "b" "d" "c" "e"
x = c("ab(A)x", "dc(B)y")
extract.pattern(x, "(\\(.*\\))") # extract the string inside brackets
## [1] "(A)" "(B)"
extract.pattern(x, "\\)(.*)") # extract the string after ")"
## [1] "x" "y"
x = c("V167.G56", "V166.R56", "V122.G41", "V163.R55", "V165.B55", "V175.R59")
extract.pattern(x, "(R|G|B)") # extract R or G or B character only.
## [1] "G" "R" "G" "R" "B" "R"
extract.pattern(x, "[RGB]{1}([0-9]+)") # extract the numbers following the R, G, B character.
## [1] "56" "56" "41" "55" "55" "59"
}
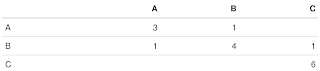
Format a confusion matrix (removing zero entry for visual clarity)
format.confusion.matrix <- function(x)
{
## Purpose: Format a confusion matrix (removing zero entry for visual clarity)
## Requires package "formattable".
## Arguments:
## x: a confusion matrix (nrow = ncol, with counts in each cell)
## Return: a formated confusion matrix in html browser and in a CSV file.
## Author: Feiming Chen, Date: 17 Apr 2017, 15:45
## ________________________________________________
x1 <- as.character(as.matrix(x))
x1[x1 == "0"] <- ""
x2 <- matrix(x1, nrow(x), ncol(x), dimnames=dimnames((x)))
write.csv(x2, file="Confusion-Matrix.csv")
formattable::formattable(as.data.frame(x2))
}
if (F) { # Unit Test
x <- matrix(c(3,1,0,1,4,0,0,1,6), 3, 3, dimnames = list(c("A", "B", "C"), c("A", "B", "C")))
format.confusion.matrix(x)
}
Sort a Data Frame
sort.df <- function(d)
{
## Purpose: Sort a Data Frame along 1st column, ties along 2nd, ..., until its last column.
## Arguments:
## d: a data frame
## Return: a sorted data frame
## Author: Feiming Chen, Date: 18 Apr 2017, 13:49
## ________________________________________________
d[ do.call(order, d), ]
}
if (F) { # Unit Test
df <- data.frame(X = c("b", "a", "a", "c"), Y = c(4, 2, 1, 3))
sort.df(df)
## X Y
## 3 a 1
## 2 a 2
## 1 b 4
## 4 c 3
sort.df(df[2:1])
## Y X
## 3 1 a
## 2 2 a
## 4 3 c
## 1 4 b
}
Thursday, April 13, 2017
Check if a vector's values are all the same
is.constant <- function(x)
{
## Purpose: Check if a vector's values are all the same.
## Arguments:
## x: a vector (numeric, character, logical, etc.)
## Return: a logical value tha is TRUE if and only if all the values are the same (constant).
## Author: Feiming Chen, Date: 13 Apr 2017, 14:59
## ________________________________________________
y <- rep.int(x[1], length(x))
isTRUE(all.equal(x, y))
}
if (F) { # Unit Test
is.constant(c(T, T, F)) # F
is.constant(c(F, F, F)) # T
is.constant(c(1, 1, 2)) # F
is.constant(c(2, 2, 2)) # T
is.constant(c("a", "a", "b")) # F
is.constant(c("b", "b", "b")) # T
}
Tuesday, April 11, 2017
Find the value that is the maximum in absolute value
my.abs.max <- function(x)
{
## Purpose: Find the value that is the maximum in absolute value
## Arguments:
## x: a numeric vector
## Return: a value in the "x" vector that is the largest in absolute value (keep the sign).
## Author: Feiming Chen, Date: 11 Apr 2017, 15:19
## ________________________________________________
i <- which.max(abs(x))
x[i]
}
if (F) { # Unit Test
my.abs.max(c(1, -3, 2)) # -3
my.abs.max(c(1, 3, -2)) # 3
}
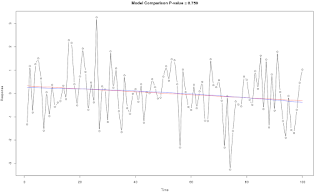
Test Time Series Second Order Curvature Significance
test.ts.2nd.order.sig <- function(x, graph=F)
{
## Purpose: Test Time Series 2nd Order Significance.
## Arguments:
## x: a numeric vector (time series)
## Return: P-value for testing the 2nd order significance.
## Author: Feiming Chen, Date: 10 Apr 2017, 14:57
## ________________________________________________
d <- data.frame(Response = x, Time = seq(x))
m1 <- lm(Response ~ Time, d)
m2 <- lm(Response ~ Time + I(Time^2), d)
a <- anova(m1, m2)
model.compare.P.value <- a[["Pr(>F)"]][2]
if (graph) {
plot(d$Time, d$Response, xlab="Time", ylab="Response", type="b")
lines(d$Time, fitted(m1), col="red")
lines(d$Time, fitted(m2), col="blue")
title(main=paste("Model Comparison P-value =", round(model.compare.P.value, 3)))
}
model.compare.P.value
}
if (F) { # Unit Test
x <- rnorm(100)
test.ts.2nd.order.sig(x, graph = T)
r <- replicate(10000, test.ts.2nd.order.sig(rnorm(20))) # should follow a uniform distribution.
## summary(r)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.00011 0.25800 0.50600 0.50500 0.75500 1.00000
}
Wednesday, April 5, 2017
Convert RGB Color Value to Spherical Coordinates
rgb2sph <- function(rgb, nbits)
{
## Purpose: Convert RGB Color Value to Spherical Coordinates
## Arguments:
## rgb: a (N X 3) matrix of RGB values (each row is a triplet of R, G, B).
## Assume each integer value range from 0 to (2^nbits - 1).
## nbits: number of bits used to code each of R, G, B channel.
## Return: a (N X 3) matrix of Shperical Coordinates (each row is a triplet of M, Theta, Phi), which are scaled to be within (0, 1).
## Author: Feiming Chen, Date: 5 Apr 2017, 10:21
## ________________________________________________
max.scale <- 2^nbits
M.scale <- sqrt(3 * max.scale^2)
angle.scale <- pi / 2
t(apply(rgb + 1, 1, function(x) {
M <- sqrt(sum(x^2)) # Color Intensity/Luminosity (0-1)
Theta <- atan(x[2] / x[1]) # Azimuthal Angle
Phi <- acos(x[3] / M) # Zenith Angle
c(M / M.scale, Theta / angle.scale, Phi / angle.scale)
}))
}
if (F) { # Unit Test
rgb <- matrix(c(0, 0, 0, 4095,4095,4095, 3527, 3513, 3504, 2470, 3034,3218), ncol=3, byrow = T)
rgb2sph(rgb, nbits = 12)
## [,1] [,2] [,3]
## [1,] 0.00024414 0.50000 0.60817
## [2,] 1.00000000 0.50000 0.60817
## [3,] 0.85832017 0.49873 0.60954
## [4,] 0.71428036 0.56498 0.56181
}
Wednesday, February 15, 2017
Look up value in a reference table (Similar to Excel's VLOOKUP function)
vlookup <- function(x, x.vector, y.vector = seq(x.vector), exact = FALSE, na.action = FALSE)
{
## PURPOSE: Look up value in a reference table (Similar to Excel's VLOOKUP function)
## ARGUMENT:
## x: a vector of Look Up Value (should not contain NA values)
## x.vector: a vector where the "x" value can be located (approximately).
## y.vector: a vector containing the return value that correponds the identified location in the "x.vector".
## Default to the index to the x.vector.
## exact: if T, the numeric values have to match exactly.
## na.action: If T and if cannot find the lookup value, return the input value.
## RETURN: a scale value that corresponds to the "x" lookup value.
## DATE: 20 Oct 2016, 16:30
## -----------------------------------------------------
ans <- NULL
if (is.numeric(x)) {
if (exact) {
ans <- y.vector[match(x, x.vector)]
} else {
ans <- y.vector[match.approx(x, x.vector)]
}
} else if (is.character(x)) {
ans <- y.vector[match(x, x.vector)]
}
if (na.action) {
idx <- is.na(ans)
ans[idx] <- x[idx]
}
ans
}
if (F) { # Unit Test
vlookup(c(68.2, 73), c(54, 60, 68.1, 72, 80), LETTERS[1:5]) # "C" "D"
vlookup(c(2, 3.1), 1:5) # 2 3
vlookup(c(2, 3.1), 1:5, exact = T) # 2 NA
vlookup(c("C", "E"), LETTERS[1:5], LETTERS[11:15]) # "M" "O"
vlookup(c("C", "X"), LETTERS[1:5], LETTERS[11:15]) # "M" NA
vlookup(c("C", "X"), LETTERS[1:5], LETTERS[11:15], na.action = T) # "M" "X"
}
match.approx <- function(x, y) {
## Purpose: Match Approximately for Numerical Data
## Arguments:
## "x": a vector of numeric values.
## "y": a vector of numeric values.
## RETURN:
## The index in "y" that indicates the closest y value to each of "x" value.
## Author: Feiming Chen, Date: 15 Feb 2017, 10:41
## ________________________________________________
sapply(x, function(x0) which.min(abs(x0 - y)))
}
if (F) {
match.approx(c(4.2, 1.2, 15), 1:10) # 4 1 10
}
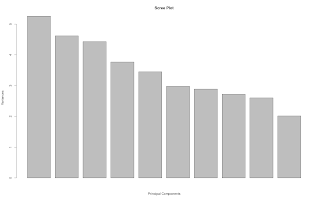
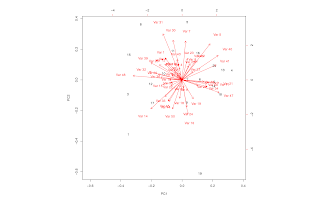
Simple Principal Component Analysis
my.pca <- function(dat.feature)
{
## Purpose: Principal Component Analysis
## Arguments:
## dat.feature: a data feature matrix, where each column is a feature vector.
## Return: PCA plots and result.
## Author: Feiming Chen, Date: 15 Feb 2017, 09:59
## ________________________________________________
p <- prcomp(dat.feature)
plot(p, main = "Scree Plot", xlab = "Principal Components")
dev.new()
biplot(p)
print(summary(p))
invisible(p)
}
if (F) { # Unit Test
d <- matrix(rnorm(1000), 20)
my.pca(d)
}
Tuesday, February 14, 2017
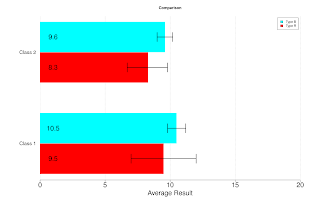
Making Barplot with Confidence Intervals
my.barplot <- function(h, hl, hu, ...)
{
## Purpose: Template function for making Barplot with Confidence Interval.
## Require R package "gplots". Refer to documentation of "gplots::barplots2".
## Arguments:
## h: Height Matrix. Each column is a group of heights. The dimnames will be used for labeling.
## hl: Lower CI. Same dimension as "h".
## hu: Upper CI. Same dimension as "h".
## ...: passed to "gplots::barplot2"
## Return:
## Author: Feiming Chen, Date: 14 Feb 2017, 14:22
## ________________________________________________
old.par <- par("mar")
par(mar = c(5, 10, 4, 5))
gplots::barplot2(h, beside = T, legend.text = T, horiz = T, col=rainbow(ncol(h)), border = NA,
cex.axis = 2, cex.names = 1.5, cex.lab=2, las = 1, xpd = F,
plot.ci = TRUE, ci.l = hl, ci.u = hu, ci.width = 0.3, ci.lwd = 2,
plot.grid = TRUE,
...) -> mp
## Put numbers on the bars
for (i in seq(ncol(mp))) text(1, mp[,i], labels=round(h[,i], 2), cex=2)
par(old.par)
}
if (F) { # Unit Test
x <- structure(c(9.5, 10.5, 1.2, 0.4, 7, 12, 9.8, 11.2, 8.3, 9.6, 0.8, 0.3, 6.7, 9.8, 9, 10.2), .Dim = c(8L, 2L), .Dimnames = list(c("Type R", "Type B", "R.SD", "B.SD", "Rl", "Ru", "Bl", "Bu"), c("Class 1", "Class 2")))
h <- x[1:2,]
hl <- x[c(5,7),]
hu <- x[c(6,8),]
my.barplot(h, hl, hu, main="Comparison", xlab="Average Result", xlim = c(0, 20))
}
Generate HTML or PNG representation of a data / table / model object
tex.print <- function(x, type=c("PNG", "HTML"), file=NULL, caption = file, digits=0) {
## Purpose: Generate HTML or PNG representation of a data/table/model object.
## Require R "memisc" package.
## Arguments:
## x: an object (table, ftable, data frame, model object, etc. that are acceptable in R "memisc" package.
## type: what to generate --
## "HTML" shows a HTML table in a web browser.
## "PNG" shows a PNG (image) file (require linux commands: latex, dvipng, display).
## file: make a copy of the PNG file with this name. Also used for the table name.
## caption: table caption.
## digits: number of decimal places to use.
## Return: Display the data representation and an image file (type="PNG") for insertion.
## Author: Feiming Chen, Date: 14 Feb 2017, 11:59
## ________________________________________________
type <- match.arg(type)
is.tabular.data <- "tabular" %in% class(x) # "x" is from the "tables" package output
is.tbl.data <- "tbl" %in% class(x) # "x" is from the "dplyr" package output
if (is.tbl.data) {
x <- as.data.frame(x)
}
switch(type,
HTML = {
## Show HTML table in a Web Browser
if (is.null(file)) {
memisc::show_html(x, digits = digits)
} else {
memisc::write_html(x, file = paste0(file,".html"), digits = digits)
}
},
PNG = {
## Generate LaTeX and PNG Image file
if (is.tabular.data) {
library(tables)
booktabs()
}
if (is.tabular.data || is.tbl.data) {
w <- Hmisc::latex(x, file="X.tex", ctable=TRUE, caption = caption, digits = digits)
if (is.tabular.data) w$style <- c("booktabs", "dcolumn")
d <- Hmisc::dvi(w) # convert to DVI file
system(paste("dvipng -q* -o X.png -T tight -D 200", d$file)) # convert to PNG file with Tight box and Resolution of 200
file.remove("X.tex")
} else {
r <- memisc:::toLatex.default(x, digits=digits, show.vars=TRUE)
f <- file("X.tex", "w")
writeLines(c("\\documentclass{article}",
"\\usepackage{booktabs}",
"\\usepackage{dcolumn}",
"\\begin{document}",
"\\pagenumbering{gobble}",
"\\begin{table}",
"\\centering"), f)
if (!is.null(caption)) writeLines(paste0("\\caption{", caption, "}"), f)
writeLines(r, f)
writeLines(c("\\end{table}", "\\end{document}"), f)
close(f)
system("latex X.tex >/dev/null") # Generate DVI file from LaTeX file (X.tex -=> X.dvi)
system("dvipng -q* -o X.png -T tight -D 200 X.dvi >/dev/null") # convert to PNG file with Tight box and Resolution of 200
file.remove("X.tex", "X.log", "X.aux", "X.dvi")
}
if (is.null(file)) {
if (!grepl("/tmp", getwd())) {
file.copy("X.png", "~/tmp/X.png", overwrite = T)
}
system("display ~/tmp/X.png &") # display the PNG file.
} else {
file.rename("X.png", paste0(file,".png"))
}
}
)
invisible(NULL)
}
if (F) { # Unit Test
x <- data.frame(age=c(1,1,1,5,5,5), height=c(10,12,9,20,23,18))
tex.print(x)
tex.print(ftable(x))
x %>% group_by(age) %>% summarise(heights = mean(height)) -> y
tex.print(y)
tex.print(x, file="Test")
tex.print(y, type="HTML")
tex.print(y, type="HTML", file = "Test")
library(tables)
a <- tabular( (Species + 1) ~ (n=1) + Format(digits=2) * (Sepal.Length + Sepal.Width) * (mean + sd), data=iris )
tex.print(a)
tex.print(a, file = "test")
}
Apply a function to a vector in an accumulative manner (like cumsum)
cum.func <- function(x, func, ...)
{
## Purpose: Apply a function to a vector in an accumulative manner (like cumsum)
## Arguments:
## x: a vector
## func: a function
## ...: passed to "func"
## Return: a vector that is the result of:
## c( func(x[1]), func(x[1:2]), func(x[1:3]), func(x[1:4]), func(x[1:5]), ... )
## Author: Feiming Chen, Date: 30 Jan 2017, 14:06
## ________________________________________________
N <- length(x)
y <- rep_len(NA, N)
for (i in 1:N) y[i] <- func(x[1:i], ...)
y[is.na(x)] <- NA
y
}
if (F) { # Unit Test
cum.func(5:1, mean) # c(5.0, 4.5, 4.0, 3.5, 3.0))
cum.func(c(NA, 5:1, NA), mean, na.rm=T) # c(NA, 5.0, 4.5, 4.0, 3.5, 3.0, NA)
cum.func(c(NA, 5:1, NA), sd, na.rm=T) # NA NA 0.70711 1.00000 1.29099 1.58114 NA
}
Overlay a new plot on an existing plot
plot.overlay <- function(..., label="2nd Y", col="blue")
{
## Purpose: Overlay a new plot on an existing plot.
## Arguments:
## ...: Passed to the "plot" function for the new plot.
## label: The secondary Y-axis label.
## col: The color of the ticks and tick labels for the new plot.
## Return: A modified plot.
## Author: Feiming Chen, Date: 6 Feb 2017, 10:19
## ________________________________________________
par(new=T)
plot(..., axes=F, xlab="", ylab="", col=col)
axis(side=4, col.ticks = col, col.axis=col)
mtext(label, side=4, line=-1, col=col)
}
if (F) { # Unit Test
plot(rnorm(100))
plot.overlay(rnorm(100), type="l", label="Test")
}
Apply a function to each consecutive pairs in a vector.
diff.func <- function(x, func)
{
## Purpose: Apply a function to each consecutive pairs in a vector.
## Arguments:
## x: a vector
## func: a function that operature on two elements (of "x")
## Return: a vector, whose length is length(x) - 1, that looks like (where N=length(x))
## func(x[2], x[1]), func(x[3], x[2]), func(x[4], x[3]), ..., func(x[N], x[N-1]).
## Author: Feiming Chen, Date: 9 Feb 2017, 11:25
## ________________________________________________
## embed in 2-dim space to make consecutive pairs
x2 <- embed(x, 2) # N - 1 pairs
## apply a function to each consecutive paris (reversely)
x3 <- apply(x2, 1, func)
x3
}
if (F) { # Unit Test
diff.func(1:5, diff) # -1 -1 -1 -1
diff.func(1:5, mean) # 1.5 2.5 3.5 4.5
}
Plot multiple lines together using "matplot"
my.line.plot <- function(x, y, ...)
{
## Purpose: Template function for making lines by group.
## Arguments:
## x: X-coords
## y: a matrix with Y-coords (each column has a set of Y values)
## ...: passed to "plot"
## Return: A plot assuming log X-axis with axes tickes given by "x".
## Author: Feiming Chen, Date: 9 Feb 2017, 14:02
## ________________________________________________n
N <- ncol(y) # number of lines
matplot(x, y, type="b", log="x", xaxt="n", lwd=2, lty=seq(N), col = rainbow(N), ...)
matpoints(x, y, log="x", xaxt="n", col = "black", ...)
axis(1, at=x)
}
if (F) { # Unit Test
x <- c(0.1, 1, 10)
y <- matrix(rnorm(9), 3)
my.line.plot(x, y, xlab="Tiime", ylab="Signal", main="Line Plot by Groups")
}
Monday, February 6, 2017
R Code: Apply a function to each element of a list (or data frame) or to each element of a list within a list of data while retaining the names attribute of each nested list
lf.compute.data.column <- function(dat.list, func, ...)
{
## Purpose: Apply a function to each column of a list (e.g. data frame). Retains the names information.
## Basis for processing more complex data (e.g. a list of a list)
## Arguments:
## dat.list: a list of data (e.g. data frame)
## func: a function that computes a vector (may have an argument "name")
## ...: passed to "func"
## Return:
## a list of outputs.
## Author: Feiming Chen, Date: 30 Jan 2017, 13:21
## ________________________________________________
this.name <- null2na(names(dat.list))
mapply(func,
dat.list,
name = this.name,
...,
SIMPLIFY = FALSE
)
}
if (F) { # Unit Test
dat.list = data.frame(x=rnorm(10), y=rnorm(10))
lf.compute.data.column(dat.list, mean)
lf.compute.data.column(dat.list, function(x, ...) { c(a=mean(x), b=sd(x)) })
lf.compute.data.column(dat.list, function(x, name) { list(a=mean(x), n=name) })
}
lf.compute.data.column.2 <- function(dat.list, func, ...)
{
## Purpose: Apply a function to a list of a list of data. Retains the names information.
## Arguments:
## dat.list: a list of a list
## func: a function that computes a vector. (may have an argument "name")
## Return: a list of list of results
## Author: Feiming Chen, Date: 6 Feb 2017, 12:50
## ________________________________________________
this.name <- null2na(names(dat.list))
mapply(
function(d, name) {
if (!is.na(name)) names(d) <- paste0(name, ".", names(d))
lf.compute.data.column(d, func)
},
dat.list,
name = this.name,
...,
SIMPLIFY = FALSE
)
}
if (F) { # Unit Test
dat.list.2 = list(test1 = data.frame(x=1:5, y=6:10), test2= data.frame(x=11:15, y=16:20))
lf.compute.data.column.2(dat.list.2, mean)
lf.compute.data.column.2(dat.list.2, function(x, ...) { c(m=mean(x), s=sd(x)) })
lf.compute.data.column.2(dat.list.2, function(x, name) { list(m=mean(x), n=name) })
}
null2na <- function(x) {
## if the value of x is NULL(as determined by length test since is.null may
## not always work), turn it into NA so that an object
## made of it will always exist.
if (length(x)==0)
x <- NA
x
}
R Code: Extract a fixed component of a list within a list and optionally apply a function
lf <- function(obj, comp, fun=identity, wrap=identity,
return.vector=TRUE, verbose=FALSE,
...) {
## WARNING: "comp" have to be a real string instead of a variable
## Functional Apply
## "comp": a character string, indicated a list component.
## "obj" is a list. "comp" is a component of each element of the
## list "obj". Apply a funcion (just an unquoted name) to each "comp" of the list
## "obj". "..." is passed to "fun". use "fun=identity" if no
## processing is needed. "wrap" is applied to the final result.
## "return.vector" will force the result to be a vector.
comp <- substitute(comp)
e <- paste(deparse(substitute(wrap)), "(lapply(obj,",
deparse(substitute(function(x) fun(x[[comp]], ...))), "))")
ans <- eval(parse(text=e))
if (verbose) cat("expression:", e, "\n")
if (return.vector)
ans <- sapply(ans, function(x) if (is.null(x)) NA else x)
ans
}
if (F) {
a <- list(x=c(1:3), y=c(4:6))
lf(a, 2) # c(x=2, y=5)
a <- list(x=list(median=3, mean=4), y=list(median=5, mean=6))
lf(a, "mean") # c(x=4, y=6)
lf(a, "mean", sqrt, verbose=T) # c(x=2, y=2.4495)
lf(a, "mean", sqrt, wrap=length, verbose=T) # 2
}
R Code: Apply a function to a vector in an accumulative manner
cum.func <- function(x, func)
{
## Purpose: Apply a function to a vector in an accumulative manner (like cumsum)
## Arguments:
## x: a vector
## func: a function
## Return: a vector that is the result of:
## c( func(x[1]), func(x[1:2]), func(x[1:3]), func(x[1:4]), func(x[1:5]), ... )
## Author: Feiming Chen, Date: 30 Jan 2017, 14:06
## ________________________________________________
N <- length(x)
y <- rep_len(NA, N)
for (i in 1:N) y[i] <- func(x[1:i])
y
}
if (F) { # Unit Test
identical(cum.func(5:1, mean), c(5.0, 4.5, 4.0, 3.5, 3.0))
}
R Function for Overlaying a New Plot on an Existing Plot
plot.overlay <- function(..., label="2nd Y", col="blue")
{
## Purpose: Overlay a new plot on an existing plot.
## Arguments:
## ...: Passed to the "plot" function for the new plot.
## label: The secondary Y-axis label.
## col: The color of the ticks and tick labels for the new plot.
## Return: A modified plot.
## Author: Feiming Chen, Date: 6 Feb 2017, 10:19
## ________________________________________________
par(new=T)
plot(..., axes=F, xlab="", ylab="", col=col)
axis(side=4, col.ticks = col, col.axis=col)
mtext(label, side=4, line=-1, col=col)
}
if (F) { # Unit Test
plot(rnorm(100))
plot.overlay(rnorm(100), type="l", label="Test")
}
Subscribe to:
Posts (Atom)