## Use Reference Semantics to pass data object to functions by reference. dat <- new.env(parent = emptyenv()) ## Put in some data dat$iris <- iris dat$cars <- cars ## Checking the original object. ls(dat) ## [1] "cars" "iris" ## An example function f <- function(d) d$N <- nrow(d$iris) + nrow(d$cars) ## Passing the data by reference: It can be modified inside the function. f(dat) ## Checking the modified object. ls(dat) ## [1] "cars" "iris" "N" dat$N ## [1] 200 identical(dat$N, nrow(dat$cars) + nrow(dat$iris)) ## [1] TRUE ## In practice, we can use f1(dat), f2(dat), f3(dat), ... ## to continuously update the information/states maintained in "dat".
Wednesday, May 30, 2018
Use R Environment Object to Share and Modify (Big) Data Within Functions
We can use the R environment object just like a list. One advantage is that it will be passed by reference to functions. So it can be modified inside functions without the need to make additional copies. Refer to Environment for more detailed explanation. Following is an example usage.
Wednesday, May 23, 2018
Examining R Profiling Data
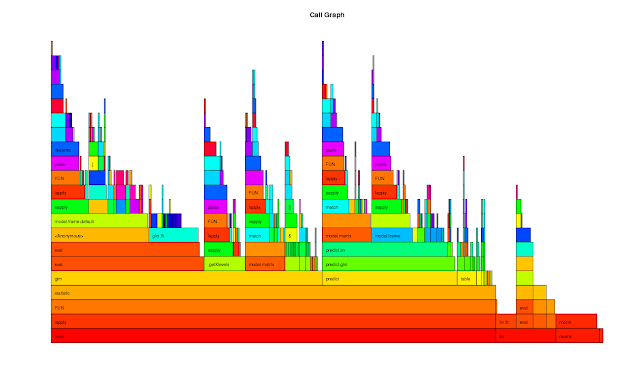
rprof <- function(srcfile) { ## Purpose: R Profiling with R "proftools" package. ## Arguments: ## srcfile: path to source file. ## Return: plots and summary of profiling. ## Author: Feiming Chen, Date: 23 May 2018, 10:31 ## ________________________________________________ library(proftools) pd0 <- profileExpr(source(srcfile)) pd <- filterProfileData(pd0, select = "withVisible", skip = 4) cat("\nSummarize by function -------------------------------------\n") print(head(funSummary(pd, srclines = FALSE), 10)) cat("\nSummarize by call -----------------------------------------\n") print(head(callSummary(pd), 10)) cat("\nHot Execution Paths ---------------------------------------\n") print(hotPaths(pd, total.pct = 10.0)) plotProfileCallGraph(pd, maxnodes = 15) flameGraph(pd) calleeTreeMap(pd) pd } if (F) { # Unit Test srcfile <- system.file("samples", "bootlmEx.R", package = "proftools") rprof(srcfile) }
The output of the Unit Test is as follows:
Summarize by function -------------------------------------
total.pct gc.pct self.pct gcself.pct
statistic 80.57 7.21 0.66 0.00
lapply 80.57 7.21 1.75 0.00
FUN 80.57 7.21 1.97 0.44
boot 80.57 7.21 0.00 0.00
glm 49.13 5.24 0.87 0.22
eval 32.75 1.97 1.97 0.22
model.frame.default 27.51 1.75 1.31 0.00
sapply 26.42 3.06 0.87 0.00
predict 24.45 1.53 0.44 0.00
predict.glm 24.02 1.53 0.44 0.00
Summarize by call -----------------------------------------
total.pct gc.pct self.pct gcself.pct
lapply -> FUN 80.57 7.21 1.09 0.22
FUN -> statistic 80.57 7.21 0.66 0.00
statistic -> glm (bootlmEx.R:33) 49.13 5.24 0.87 0.22
boot (bootlmEx.R:39) -> lapply 38.21 3.93 0.00 0.00
boot (bootlmEx.R:44) -> lapply 37.55 3.06 0.00 0.00
eval -> eval 30.13 1.97 0.87 0.22
glm (bootlmEx.R:33) -> eval 27.73 1.75 0.87 0.00
statistic -> predict (bootlmEx.R:35) 24.45 1.53 0.44 0.00
predict (bootlmEx.R:35) -> predict.glm 24.02 1.53 0.44 0.00
sapply -> lapply 23.14 3.06 1.75 0.00
Hot Execution Paths ---------------------------------------
path total.pct self.pct
boot (bootlmEx.R:39) 38.21 0.00
. lapply 38.21 0.00
. . FUN 38.21 0.00
. . . statistic 38.21 0.00
. . . . glm (bootlmEx.R:33) 24.67 0.44
. . . . . eval 13.76 0.44
. . . . . . eval 13.32 0.22
. . . . predict (bootlmEx.R:35) 12.23 0.00
. . . . . predict.glm 12.01 0.44
. . . . . . predict.lm 11.35 0.22
boot (bootlmEx.R:44) 37.55 0.00
. lapply 37.55 0.00
. . FUN 37.55 0.00
. . . statistic 37.55 0.00
. . . . glm (bootlmEx.R:33) 24.45 0.44
. . . . . eval 13.97 0.44
. . . . . . eval 13.54 0.00
. . . . predict (bootlmEx.R:35) 12.23 0.22
. . . . . predict.glm 12.01 0.00 . . . . . . predict.lm 11.35 0.44 lm (bootlmEx.R:52) 10.92 0.00
Monday, May 21, 2018
Predict with a Classification Tree ("rpart") Model
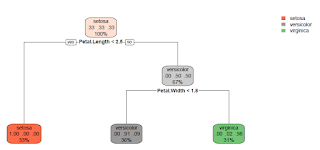
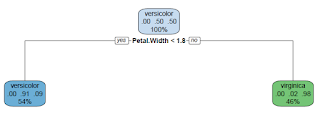
report.model.rpart <- function(fmla, dat, main = "Classification Tree") { ## Purpose: Report a Classification Tree ("rpart") model and make plots. ## Assume the response is a categorical variable (factor). ## Require package "rpart" and "rpart.plot". ## Arguments: ## fmla: formula for the predictive model ## dat: data frame for the training data ## main: Title for the plot ## Return: ## Author: Feiming Chen, Date: 18 May 2018, 11:41 ## ________________________________________________ library(rpart) library(rpart.plot) ans <- rpart(fmla, dat) rpart.plot(ans, main = main) mf <- model.frame(fmla, dat) Response <- factor(model.response(mf)) # extract response Predicted.Response <- factor(predict(ans, type = "class")) err.mis(Response, Predicted.Response, file="Tree-Model-CART") if (nlevels(Response) == 2) { H0.label <- levels(Response)[1] o1 <- sens.spec(Response, Predicted.Response, H0=H0.label) print(o1) o2 <- sens.spec(Response, Predicted.Response, H0=H0.label, get.ppv = TRUE) print(o2) } ans } if (F) { # Unit Test data(iris) report.model.rpart(Species ~ ., iris) # Predict 3 species report.model.rpart(Species ~ ., subset(iris, Species != "setosa")) # Predict 2 species }
Unit Test 1: Predict 3 Species
Unit Test 2: Predict 2 Species
Monday, May 14, 2018
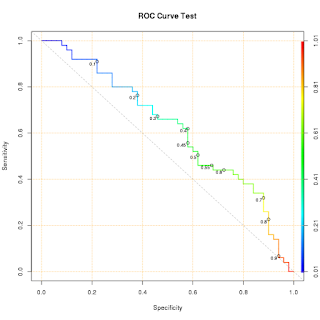
Make a ROC plot with possible cutoff points
roc.curve <- function(response, predicted, cutoff = c(seq(0.1, 0.9, 0.1), 0.45, 0.55), ...) { ## Purpose: Make a ROC plot with possible cutoff points. ## Require R package "ROCR" ## Arguments: ## response: a vector of truth (0/FALSE/"negative" or 1/TRUE/"positive") ## predicted: a vector of prediction (continuous); ## cutoff: a list of values to be plotted on ROC curve. ## ...: passed to "plot". ## Return: a ROC plot. ## Author: Feiming Chen, Date: 14 May 2018, 15:01 ## ________________________________________________ require(ROCR) ans <- ROCR::prediction(predicted, response) ## ROC for Sensitivity vs. Specificity. plot((pp <- ROCR::performance(ans, "sens", "spec")), colorize=T, print.cutoffs.at=cutoff, text.adj=c(1.2, 1.2), text.cex=0.7, lwd=2, ...) grid(col="orange") ## Draw a "line of no-discrimination". ## Sens = P(X=+ | T=+), Spec = P(X=- | T=-), ## if X is independent of T, then Sens + Spec = P(X+)+P(X-) = 1, so the pair ## (Sens, Spec) lies on a off-diagonal line. abline(c(1, -1), col="gray70", lty=2) return(invisible(pp)) } if (F) { roc.curve(rep(c(0,1), 50), runif(100), main = "ROC Curve Test") }
Tuesday, May 8, 2018
Calculate the sample size required for detecting at least one rare event
sample.size.for.rare.event.detection <- function(alpha = 0.05, M = 0.01) { ## Purpose: Calculate the sample size required for detecting at least one rare event. ## Arguments: ## alpha: Type I Error. Sensitivity = 1 - alpha. ## M: Probability for the rare event. Default to 1%. ## Return: A minimum sample size required for the detection of rare event ## with (1-alpha) probability (aka. confidence). ## Author: Feiming Chen, Date: 8 May 2018, 10:36 ## ________________________________________________ N <- ceiling(log(alpha) / log(1 - M)) N } if (F) { # Unit Test sample.size.for.rare.event.detection() # 299 ## Detecting 0.5% event with 99% confidence sample.size.for.rare.event.detection(alpha = 0.01, M = 0.005) # 919 }
Below is an application example.
Tuesday, May 1, 2018
Calculate the entropy of a signal emulating the classic Shannon definition.
signal.entropy <- function(x) { ## Purpose: Calculate the entropy of a signal, emulating the classic Shannon definition. ## Arguments: ## x: A signal vector ## Return: An entropy number ## Author: Feiming Chen, Date: 1 May 2018, 13:07 ## ________________________________________________ x2 <- x^2 tot <- sum(x2) if (tot == 0) { H <- NA } else { p <- x2 / tot H <- - sum(p * log10(p)) } H } if (F) { # Unit Test x <- rnorm(100) signal.entropy(x) signal.entropy(rep(1,100)) # 2 signal.entropy(rep(1,10000)) # 4 }
Subscribe to:
Posts (Atom)