feature.filtering.ns <- function(X, df = 10, graph = FALSE)
{
## Purpose: Feature Filtering with Natural B-Splines for Functional Modeling Application.
## Preprocess the feature data (design matrix) to enforce a natural regularization.
## In other words, project each observation to each of "df" axis represented by B-spline Basis.
## Arguments:
## X: a design matrix (predictors) of dimension N x p (N observations, p predictors).
## If "X" is a vector, convert it to a matrix with one row.
## df: Degrees of Freedom (number of natural B-splines basis functions to approximate the (continuous) coefficient function.
## Beta (p x 1) = H (p x df) Theta (df x 1), where H represents "df" number of continuous basis functions
## in the coefficient space, and Theta represents the linear combination parameters for constructing Beta.
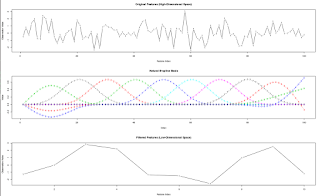
## graph: if TRUE, plot each observation (each row of X) as a time series,
## its projection to the reduced space (formed by the natural B-spline basis),
## and the natural B-spline basis.
## Return: Preprocessed Features (X H) of dimension N x df, which can be used as "df" input into a predictive model.
## X Beta = X H Theta = (X H) Theta
## Author: Feiming Chen, Date: 3 May 2017, 12:12
## ________________________________________________
if (is.vector(X)) X <- matrix(X, nrow = 1)
p <- ncol(X) # number of predictors
require(splines)
## B-splines for Natural Cubic Spline: a p x df matrix.
H <- ns(1:p, df=df)
## Filtered Features. Dimensions: N x p p x df = N x df
## Project each observation "x" into one of "df" Basis (each column of H matrix) so that
## each "x" is represented by "df" coefficient (coordinates in the coordinate system defined by "H")
ans <- X %*% H
if (graph) {
par(mfrow=c(3,1))
matplot(t(X), type = "b", xlab = "Feature Index", ylab = "Observation Value",
main = "Original Features (High-Dimensional Space)")
matplot(H, type="b", xlab = "Index", ylab ="Value", main="Natural B-spline Basis")
matplot(t(ans), type = "b", xlab = "Feature Index", ylab = "Observation Value",
main = "Filtered Features (Low-Dimensional Space)")
par(mfrow=c(1,1))
}
ans
}
if (F) { # Unit Test
X <- matrix(rnorm(1000), 20, 50)
y <- feature.filtering.ns(X)
y <- feature.filtering.ns(X, df = 20)
x <- rnorm(100)
y <- feature.filtering.ns(x, graph=T)
}