calc.reference.interval <- function(val, wgt) { ## Purpose: Calculate Nonparametric 95% Reference Interval. ## Notes: ## 1) Each Reference Interval needs at least 120 reference observations (per CLSI C28-A3c). ## 2) A general criterion for determining sample size is that the width of the 90% confidence interval (CI) ## for a reference limit should be acceptably small relative to the width of the 95% reference interval ## itself (Harris and Boyd); these authors recommend that the width of the 90% CI be less than 0.2 times ## the width of the reference interval. ## Arguments: ## val: numerical measurements ## wgt: weight (frequency) of these measurements ## Return: a Nonparametric 95% Reference Interval. ## Author: Feiming Chen ## ________________________________________________ x <- rep(val, wgt) cat("N =", length(x), "\n") cat("Reference Interval:\n") print(q <- quantile(x, probs = c(0.025, 0.975))) ## Confidence Intervals for the Nonparametric Method (Using Bootstrap with 2000 Reps) require(boot) b <- boot(x, function(x, i) { quantile(x[i], probs = c(0.025, 0.975)) }, R = 2000) cat("\n90% Bootstrap CI for Lower 95% Reference Limit:") L <- boot.ci(b, conf=0.90, type="basic", index=1) print(L2 <- L$basic[1,c(4,5)]) cat("\n90% Bootstrap CI for Upper 95% Reference Limit:") U <- boot.ci(b, conf=0.90, type="basic", index=2) print(U2 <- U$basic[1,c(4,5)]) cat("\nRatio of Limit CI Width to Reference Interval Width (Recommend: < 0.2):\n") q2 <- diff(q) # Width of Reference Interval L3 <- diff(L2) # Width of Lower Limit 90% CI U3 <- diff(U2) # Width of Upper Limit 90% CI cat("Lower Limit CI Width Ratio:", round(L3 / q2, 3), "\n") cat("Upper Limit CI Width Ratio:", round(U3 / q2, 3), "\n") list(Reference.Limit = q, Lower.Limit.CI = L2, Upper.Limit.CI = U2) } if (F) { # Unit Test ## Data from CLSI C28-A3c (Reference Interval) Table 4 (Frequency Distributions of Calcium in 240 Medical Students by Sex) ## Only Women's Data. CLSI Reported "Nonparametric" Method: N = 120, Calcium.Women = (8.9, 10.2), ## Lower Limit 90% CI = (8.8, 9.1), Upper Limit 90% CI = (10.1, 10.3) val <- c(8.8,8.9,9.0,9.1,9.2,9.3,9.4,9.5,9.6,9.7,9.8,9.9,10.0,10.1,10.2,10.3,10.4,10.5,10.6) wgt <- c(1,2,1,3,11,11,8,16,16,26,8,7,3,2,3,2,0,0,0) ans <- calc.reference.interval(val, wgt) ## 2.5% 97.5% ## 8.9975 10.2000 }
Friday, December 7, 2018
Calculate Nonparametric 95% Reference Interval
Friday, November 30, 2018
Variance Component Analysis for One-Way Random Model
var.comp.one.way.model <- function(fmla, dat) { ## Purpose: Variance Component Analysis for One-Way Random Model. ## Application: Repeatability and Reproducibility Analysis (R&R for Precision Study). ## Use Method of Moment estimation. ## Arguments: ## fmla: a formula with one response and one factor (One-way Random Model). ## dat: a data frame ## Return: Variance Component Analysis such as ## DF: Degrees of Freedom ## VC: Variance Component (with proportion of total in percentage) ## SD: Standard Deviation (from the squared root of Variance Component) ## CV%: Coefficient of Variation as Percentage ## Author: Feiming Chen ## ________________________________________________ a <- aov(fmla, dat) b <- summary(a) cat("Analysis of Variance:\n") print(b) # Analysis of Variance MEAN <- mean(a$model[[1]]) # Mean N <- nrow(a$model) # Number of observations cat("\nMean: ", round(MEAN, 3), " (N =", N, ")\n", sep="") d <- b[[1]] VC.error <- d$"Mean Sq"[2] # Mean Sum of Squares for Error = Variance Component for Error (Repeatability) Q2 <- d$"Sum Sq"[1] # Sum of Squares for Treatment DF.treatment <- d$Df[1] # DF for Treatment N.i <- table(a$model[[2]]) # Number of Replicates in Each Treatment Choice VC.treatment <- (Q2 - DF.treatment * VC.error) / (N - sum(N.i^2) / N) VC <- c(VC.treatment, VC.error, VC.treatment + VC.error) VC.pct.total <- VC / VC[3] * 100 SD <- sqrt(VC) CV.pct <- SD / MEAN * 100 ans <- data.frame(Name = c(attr(a$terms, "term.labels"), "Error", "Total"), VC = round(VC, 3), VC.Pct.Total = round(VC.pct.total, 3), SD = round(SD, 3), CV.Pct = round(CV.pct, 3)) cat("\nVariance Component Analysis:\n") ans } if (F) { # Unit Test dat <- data.frame(Yield=c(3.9, 4.05, 4.25, 3.60, 4.2, 4.05, 3.85, 4.15, 4.6, 4.15, 4.4, 3.35, 3.8), Variety=factor(c(1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4))) fmla <- Yield ~ Variety var.comp.one.way.model(fmla, dat) ## Validation library(VCA) anovaVCA(fmla, dat) }
Friday, June 1, 2018
Compare the speed of product vs exp/sum/log calculations
## Compare the speed of product vs exp/sum/log calculations library(microbenchmark) x <- runif(1000) + 0.5 all.equal(prod(x), exp(sum(log(x)))) ## [1] TRUE microbenchmark::microbenchmark( prod(x), exp(sum(log(x))) ) ## Unit: microseconds ## expr min lq mean median uq max neval ## prod(x) 2.075 2.107 2.52904 2.1945 2.5285 13.964 100 ## exp(sum(log(x))) 51.525 52.075 62.89344 52.6135 62.5120 400.676 100
We can see it is much faster to use the straightforward calculation of "prod".
Wednesday, May 30, 2018
Use R Environment Object to Share and Modify (Big) Data Within Functions
We can use the R environment object just like a list. One advantage is that it will be passed by reference to functions. So it can be modified inside functions without the need to make additional copies. Refer to Environment for more detailed explanation. Following is an example usage.
## Use Reference Semantics to pass data object to functions by reference. dat <- new.env(parent = emptyenv()) ## Put in some data dat$iris <- iris dat$cars <- cars ## Checking the original object. ls(dat) ## [1] "cars" "iris" ## An example function f <- function(d) d$N <- nrow(d$iris) + nrow(d$cars) ## Passing the data by reference: It can be modified inside the function. f(dat) ## Checking the modified object. ls(dat) ## [1] "cars" "iris" "N" dat$N ## [1] 200 identical(dat$N, nrow(dat$cars) + nrow(dat$iris)) ## [1] TRUE ## In practice, we can use f1(dat), f2(dat), f3(dat), ... ## to continuously update the information/states maintained in "dat".
Wednesday, May 23, 2018
Examining R Profiling Data
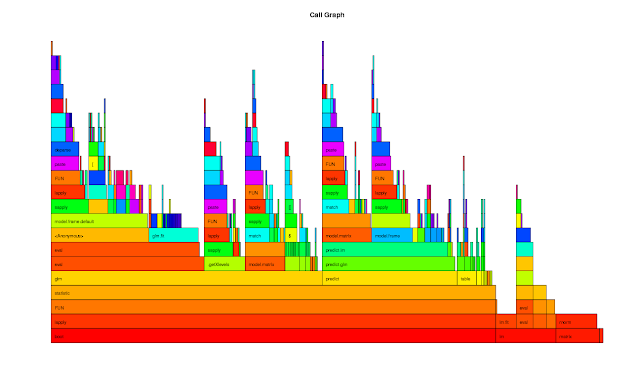
rprof <- function(srcfile) { ## Purpose: R Profiling with R "proftools" package. ## Arguments: ## srcfile: path to source file. ## Return: plots and summary of profiling. ## Author: Feiming Chen, Date: 23 May 2018, 10:31 ## ________________________________________________ library(proftools) pd0 <- profileExpr(source(srcfile)) pd <- filterProfileData(pd0, select = "withVisible", skip = 4) cat("\nSummarize by function -------------------------------------\n") print(head(funSummary(pd, srclines = FALSE), 10)) cat("\nSummarize by call -----------------------------------------\n") print(head(callSummary(pd), 10)) cat("\nHot Execution Paths ---------------------------------------\n") print(hotPaths(pd, total.pct = 10.0)) plotProfileCallGraph(pd, maxnodes = 15) flameGraph(pd) calleeTreeMap(pd) pd } if (F) { # Unit Test srcfile <- system.file("samples", "bootlmEx.R", package = "proftools") rprof(srcfile) }
The output of the Unit Test is as follows:
Summarize by function -------------------------------------
total.pct gc.pct self.pct gcself.pct
statistic 80.57 7.21 0.66 0.00
lapply 80.57 7.21 1.75 0.00
FUN 80.57 7.21 1.97 0.44
boot 80.57 7.21 0.00 0.00
glm 49.13 5.24 0.87 0.22
eval 32.75 1.97 1.97 0.22
model.frame.default 27.51 1.75 1.31 0.00
sapply 26.42 3.06 0.87 0.00
predict 24.45 1.53 0.44 0.00
predict.glm 24.02 1.53 0.44 0.00
Summarize by call -----------------------------------------
total.pct gc.pct self.pct gcself.pct
lapply -> FUN 80.57 7.21 1.09 0.22
FUN -> statistic 80.57 7.21 0.66 0.00
statistic -> glm (bootlmEx.R:33) 49.13 5.24 0.87 0.22
boot (bootlmEx.R:39) -> lapply 38.21 3.93 0.00 0.00
boot (bootlmEx.R:44) -> lapply 37.55 3.06 0.00 0.00
eval -> eval 30.13 1.97 0.87 0.22
glm (bootlmEx.R:33) -> eval 27.73 1.75 0.87 0.00
statistic -> predict (bootlmEx.R:35) 24.45 1.53 0.44 0.00
predict (bootlmEx.R:35) -> predict.glm 24.02 1.53 0.44 0.00
sapply -> lapply 23.14 3.06 1.75 0.00
Hot Execution Paths ---------------------------------------
path total.pct self.pct
boot (bootlmEx.R:39) 38.21 0.00
. lapply 38.21 0.00
. . FUN 38.21 0.00
. . . statistic 38.21 0.00
. . . . glm (bootlmEx.R:33) 24.67 0.44
. . . . . eval 13.76 0.44
. . . . . . eval 13.32 0.22
. . . . predict (bootlmEx.R:35) 12.23 0.00
. . . . . predict.glm 12.01 0.44
. . . . . . predict.lm 11.35 0.22
boot (bootlmEx.R:44) 37.55 0.00
. lapply 37.55 0.00
. . FUN 37.55 0.00
. . . statistic 37.55 0.00
. . . . glm (bootlmEx.R:33) 24.45 0.44
. . . . . eval 13.97 0.44
. . . . . . eval 13.54 0.00
. . . . predict (bootlmEx.R:35) 12.23 0.22
. . . . . predict.glm 12.01 0.00 . . . . . . predict.lm 11.35 0.44 lm (bootlmEx.R:52) 10.92 0.00
Monday, May 21, 2018
Predict with a Classification Tree ("rpart") Model
report.model.rpart <- function(fmla, dat, main = "Classification Tree") { ## Purpose: Report a Classification Tree ("rpart") model and make plots. ## Assume the response is a categorical variable (factor). ## Require package "rpart" and "rpart.plot". ## Arguments: ## fmla: formula for the predictive model ## dat: data frame for the training data ## main: Title for the plot ## Return: ## Author: Feiming Chen, Date: 18 May 2018, 11:41 ## ________________________________________________ library(rpart) library(rpart.plot) ans <- rpart(fmla, dat) rpart.plot(ans, main = main) mf <- model.frame(fmla, dat) Response <- factor(model.response(mf)) # extract response Predicted.Response <- factor(predict(ans, type = "class")) err.mis(Response, Predicted.Response, file="Tree-Model-CART") if (nlevels(Response) == 2) { H0.label <- levels(Response)[1] o1 <- sens.spec(Response, Predicted.Response, H0=H0.label) print(o1) o2 <- sens.spec(Response, Predicted.Response, H0=H0.label, get.ppv = TRUE) print(o2) } ans } if (F) { # Unit Test data(iris) report.model.rpart(Species ~ ., iris) # Predict 3 species report.model.rpart(Species ~ ., subset(iris, Species != "setosa")) # Predict 2 species }
Unit Test 1: Predict 3 Species
Unit Test 2: Predict 2 Species
Monday, May 14, 2018
Make a ROC plot with possible cutoff points
roc.curve <- function(response, predicted, cutoff = c(seq(0.1, 0.9, 0.1), 0.45, 0.55), ...) { ## Purpose: Make a ROC plot with possible cutoff points. ## Require R package "ROCR" ## Arguments: ## response: a vector of truth (0/FALSE/"negative" or 1/TRUE/"positive") ## predicted: a vector of prediction (continuous); ## cutoff: a list of values to be plotted on ROC curve. ## ...: passed to "plot". ## Return: a ROC plot. ## Author: Feiming Chen, Date: 14 May 2018, 15:01 ## ________________________________________________ require(ROCR) ans <- ROCR::prediction(predicted, response) ## ROC for Sensitivity vs. Specificity. plot((pp <- ROCR::performance(ans, "sens", "spec")), colorize=T, print.cutoffs.at=cutoff, text.adj=c(1.2, 1.2), text.cex=0.7, lwd=2, ...) grid(col="orange") ## Draw a "line of no-discrimination". ## Sens = P(X=+ | T=+), Spec = P(X=- | T=-), ## if X is independent of T, then Sens + Spec = P(X+)+P(X-) = 1, so the pair ## (Sens, Spec) lies on a off-diagonal line. abline(c(1, -1), col="gray70", lty=2) return(invisible(pp)) } if (F) { roc.curve(rep(c(0,1), 50), runif(100), main = "ROC Curve Test") }
Tuesday, May 8, 2018
Calculate the sample size required for detecting at least one rare event
sample.size.for.rare.event.detection <- function(alpha = 0.05, M = 0.01) { ## Purpose: Calculate the sample size required for detecting at least one rare event. ## Arguments: ## alpha: Type I Error. Sensitivity = 1 - alpha. ## M: Probability for the rare event. Default to 1%. ## Return: A minimum sample size required for the detection of rare event ## with (1-alpha) probability (aka. confidence). ## Author: Feiming Chen, Date: 8 May 2018, 10:36 ## ________________________________________________ N <- ceiling(log(alpha) / log(1 - M)) N } if (F) { # Unit Test sample.size.for.rare.event.detection() # 299 ## Detecting 0.5% event with 99% confidence sample.size.for.rare.event.detection(alpha = 0.01, M = 0.005) # 919 }
Below is an application example.
Tuesday, May 1, 2018
Calculate the entropy of a signal emulating the classic Shannon definition.
signal.entropy <- function(x) { ## Purpose: Calculate the entropy of a signal, emulating the classic Shannon definition. ## Arguments: ## x: A signal vector ## Return: An entropy number ## Author: Feiming Chen, Date: 1 May 2018, 13:07 ## ________________________________________________ x2 <- x^2 tot <- sum(x2) if (tot == 0) { H <- NA } else { p <- x2 / tot H <- - sum(p * log10(p)) } H } if (F) { # Unit Test x <- rnorm(100) signal.entropy(x) signal.entropy(rep(1,100)) # 2 signal.entropy(rep(1,10000)) # 4 }
Wednesday, April 25, 2018
Calibration Plot for Probability Prediction
calibration.plot <- function (y, p, main="Calibration Plot") { ## Purpose: Calibration Plot for Probability Forecast ## Arguments: ## y: the observed binary outcomes (0-1 response). ## p: the probability forecasts (estimating E(Y|X)). ## Return: ## - Calibration Plot ## - A (Calibration-in-the-Large), and B (Calibration-Slope) ## Author: Feiming Chen ## ________________________________________________ ## Fit Non-parametric model newp <- seq(0, 1, length=100) yy <- predict(loess(y ~ p, span=1), newp, se=T) # LOESS Smoothing yy.ok <- !is.na(yy$fit) yy$fit <- yy$fit[yy.ok] # calibration curve yy$se.fit <- yy$se.fit[yy.ok] # standard error newp <- newp[yy.ok] se.lower <- yy$fit - 2 * yy$se.fit # confidence band (lower limit) se.upper <- yy$fit + 2 * yy$se.fit # confidence band (upper limit) ## Fit logist regression and obtain the intercept and slope dat <- data.frame(y = y, x = log(p / (1 - p))) f <- glm(y ~ x, family = binomial, data = dat) # "Cox, 1958, Biometrika" ce <- round(coef(f), 2) A <- ce[1] # A = Intercept B <- ce[2] # B = Slope cf <- round(confint(f), 2) # 95% CI par(pty="s") plot(c(0,1), c(0,1), type="n", xlab="Predicted Probability", ylab="Observed Frequencies", xaxs="i", yaxs="i", las=1, main=main) polygon(c(newp, rev(newp), newp[1]), # plot confidence band c(se.lower, rev(se.upper), se.lower[1]), col = "gray", border = NA) rug(p[y == 0], side=1, col="navy") # binary response 0's rug(p[y == 1], side=3, col="navy") # binary response 1's abline(0, 1, col="red") # perfect calibration line abline(h=0.5, col="red", lty=2) # perfect irrelevance (no-discriminatino) line abline(v=0.5, lty=2) # default cut-off at 50% for call lines(newp, yy$fit, lwd=2, col="blue") # plot calibration curve text(0.2, 0.8, labels = paste("A = ", A, "(", cf[1,1], ",", cf[1,2], ")")) text(0.2, 0.77, labels = paste("B = ",B, "(", cf[2,1], ",", cf[2,2], ")")) par(pty="m") } if (F) { # Unit Test calibration.plot(y = rbinom(1000, 1, 0.5), p = runif(1000), main = "Random Prediction") p <- runif(1000) y <- sapply(p, function(x) rbinom(1, size = 1, prob = x)) # perfect probability prediction calibration.plot(y, p, main = "Perfect Prediction") }
R Analysis Report with Emacs Org Mode
Write a test.org file as follows:
Export to HTML file (1st image) with C-c C-e h o
Export to PDF file (2nd image) with C-c C-e l o

* Test You can write inline expressions like \pi = src_R{pi}. Here is a code chunk. #+NAME:foo #+begin_src R :file plot.png :exports both :results output graphics x <- rnorm(100) summary(x) plot(x) #+end_src
Export to HTML file (1st image) with C-c C-e h o
Export to PDF file (2nd image) with C-c C-e l o

R Analysis Report with Sweave/Knitr + Emacs + AUCTEX
- In Emacs, compose a file "test.Rnw".
- Enter command for knitting: "M-n r" (Noweb => Sweaving/Tangling => Knit)
- Enter command for generating PDF report: "M-n P".
\documentclass{article} \title{Test} \begin{document} You can write inline expressions like $\pi=\Sexpr{pi}$. Here is a code chunk. <<a>>= x <- rnorm(100) summary(x) plot(x) @ \end{document}
Monday, January 15, 2018
Equivalence Test for Two Means by TOST (Two One-Sided Test) Procedure
mean.equiv.tost.test <- function(x, y, margin=1, alpha=0.05) {
## Equivalence Test for Two Means by TOST (Two One-Sided Test) Procedure
## Assumption: Normality.
## Reference: Understanding Equivalence and Noninferiority Testing (Walker and Nowacki)
## INPUT:
## x: a vector of continuous scale.
## y: another vector of continuous scale (to compare with x)
## margin: Equivalence Margin (defaul to 1)
## alpha: Significance Level.
cl <- 1 - 2 * alpha
a <- t.test(x, y, conf.level=cl)
ci <- a$conf.int
print(a)
cat("Equivalence Test for Two Means by TOST Procedure:\n")
cat("Equivalence Margin =", margin, "\n")
if (ci[1] >= -margin && ci[2] <= margin) {
cat("Equivalence Established! (Significance Level =", 100*alpha, "%)\n\n")
} else cat("Equivalence NOT Established. (Significance Level =", 100*alpha, "%)\n\n")
plot(0, 0, xlim=c(-2*margin, 2*margin), ylim=c(-1,1), axes=F, xlab="Difference in Efficacies", ylab="", type="n")
axis(side=1, at=c(-2*margin, -margin, 0, margin, 2*margin), labels=c("", paste("-", margin), "0", margin, ""))
abline(v=c(-margin, 0, margin), lty=c(2,1,2))
lines(ci, c(0,0), lwd=3)
lines(rep(ci[1], 2), c(-0.1, 0.1), lwd=2)
lines(rep(ci[2], 2), c(-0.1, 0.1), lwd=2)
lines(rep(-diff(a$estimate), 2), c(-0.1, 0.1), lwd=4)
}
if (F) {
mean.equiv.tost.test(rnorm(20), rnorm(20, mean=0.3))
}
Equivalence Test for Proportions by TOST (Two One-Sided Test) Procedure
prop.equiv.tost.test <- function(x, n, margin=0.2, alpha=0.05) {
## Equivalence Test for Proportions by TOST (Two One-Sided Test) Procedure
## Reference: Understanding Equivalence and Noninferiority Testing (Walker and Nowacki)
## INPUT:
## x: a vector of two success counts,
## n: a vector of two sample sizes
## margin: Equivalence Margin (defaul to 20%)
## alpha: Significance Level.
cl <- 1 - 2 * alpha
a <- prop.test(x, n, conf.level= cl)
## Using Score Interval for Difference of Proportions
require(PropCIs)
b <- diffscoreci(x[1], n[1], x[2], n[2], conf.level= cl)
ci <- b$conf.int
cat("\nEquivalence Test for Proportions by TOST Procedure:\n")
cat("1st Trial =", x[1], "/", n[1], "=", round(x[1]/n[1]*100, 1), "% , 2nd Trial =", x[2], "/", n[2], "=", round(x[2]/n[2]*100, 1), "%\n")
pdiff <- -diff(a$estimate) # p1 - p2
cat("Difference in Efficacies =", 100 * round(pdiff, 3), "%\n")
cat(100*cl, "% Confidence Interval = [", 100*round(ci[1], 3), "% ,", 100*round(ci[2], 3), "% ]\n")
cat("Equivalence Margin =", 100*margin, "%\n")
if (ci[1] >= -margin && ci[2] <= margin) {
cat("Equivalence Established! (Significance Level =", 100*alpha, "%)\n\n")
} else cat("Equivalence NOT Established. (Significance Level =", 100*alpha, "%)\n\n")
cat("(Assumption: 1st Trial is New, 2nd one is Current (Reference), and Higher Proportion is Better.)\n")
if (ci[1] >= -margin) {
cat("Noninferiority Established! (Significance Level =", 100*alpha, "%)\n\n")
} else cat("Noninferiority NOT Established. (Significance Level =", 100*alpha, "%)\n\n")
## Sample Size Determination (TOST)
p <- x/n
c1 <- ((qnorm(0.95) + qnorm(0.8)))^2 # Type I Error = 5%, Type II Error = 20%, Power = 80%
pd <- abs(pdiff)
if (pd >= margin) N <- NA else N <- ceiling(c1 * (p[1]*(1-p[1])+p[2]*(1-p[2])) / (pd - margin)^2 )
cat("Sample Size Required (using only input proportions and margin) =", N, "(Power = 80 % )\n")
plot(0, 0, xlim=c(-2*margin, 2*margin), ylim=c(-1,1), axes=F, xlab="Difference in Efficacies", ylab="", type="n")
axis(side=1, at=c(-2*margin, -margin, 0, margin, 2*margin), labels=c("", paste("-",
100*margin, "%"), "0",
paste(100*margin, "%"), ""))
abline(v=c(-margin, 0, margin), lty=c(2,1,2))
lines(ci, c(0,0), lwd=3)
lines(rep(ci[1], 2), c(-0.1, 0.1), lwd=2)
lines(rep(ci[2], 2), c(-0.1, 0.1), lwd=2)
lines(rep(pdiff, 2), c(-0.1, 0.1), lwd=4)
}
if (F) {
prop.equiv.tost.test(c(80, 90), c(100,100))
prop.equiv.tost.test(c(143, 144), c(281,281), margin=0.12, alpha=0.025)
}
Wednesday, January 3, 2018
Replace NA value with the last numeric value in each row
replace.NA.with.last.value.in.each.row <- function(dat)
{
## Purpose: Replace NA value with the last numeric value in each row.
## Arguments:
## dat: a data frame that may have NA in the tail of each row.
## Return: an updated data frame with no NA values.
## Author: Feiming Chen, Date: 3 Jan 2018, 11:34
## ________________________________________________
last.value <- apply(dat, 1, function(x) as.numeric(tail(na.omit(x), 1))) # last numeric value in each row.
n <- is.na(dat)
for (i in 1:nrow(dat)) {
dat[i, n[i,]] <- last.value[i]
}
dat
}
if (F) { # Unit Test
dat <- data.frame(ID=LETTERS[1:5], V1=c(1:4, NA), V2=c(6:7, NA, NA, NA), V3 = c(8, rep(NA, 4)))
## ID V1 V2 V3
## 1 A 1 6 8
## 2 B 2 7 NA
## 3 C 3 NA NA
## 4 D 4 NA NA
## 5 E NA NA NA
replace.NA.with.last.value.in.each.row(dat)
## ID V1 V2 V3
## 1 A 1 6 8
## 2 B 2 7 7
## 3 C 3 3 3
## 4 D 4 4 4
## 5 E NA NA NA
}
Make a combined data frame with a source label using the list names
rbind.df.with.list.names <- function(obj)
{
## Purpose: Make a combined data frame with a source label using the list names.
## Arguments:
## obj: It is a list of data frames to be "rbind" together.
## Return: a single combined data frame with all the sub data frames in "obj"
## with an addtional column "SOURCE0" from the names of the list "obj",
## and another addtional column "SOURCE1" that abbreviates "SOURCE0".
## Author: Feiming Chen, Date: 2 Jan 2018, 11:00
## ________________________________________________
o <- do.call(rbind, obj)
o$SOURCE0 <- rep(names(obj), sapply(obj, nrow))
o$SOURCE1 <- abbreviate(o$SOURCE0)
o
}
if (F) { # Unit Test
obj <- list(A = data.frame(x = 1:3), B = data.frame(x = 4:6))
rbind.df.with.list.names(obj)
## x SOURCE0 SOURCE1
## A.1 1 A A
## A.2 2 A A
## A.3 3 A A
## B.1 4 B B
## B.2 5 B B
## B.3 6 B B
}
Subscribe to:
Comments (Atom)